1 项目概述
| 项目编号 | GDMR21010245_sup_6 |
| 样品信息 | LL-1¦LL-2¦LL-3¦LN-1¦LN-2¦LN-3
TL-1¦TL-2¦TL-3¦TN-1¦TN-2¦TN-3 |
| 分组方案 | LL :LL-1&LL-2&LL-3¦LN :LN-1&LN-2&LN-3¦TL :TL-1&TL-2&TL-3¦TN :TN-1&TN-2&TN-3 |
| 两组差异分析方案 | LN-vs-LL¦TN-vs-TL¦LL-vs-TL¦LN-vs-TN |
2 项目简介
2.1 背景介绍
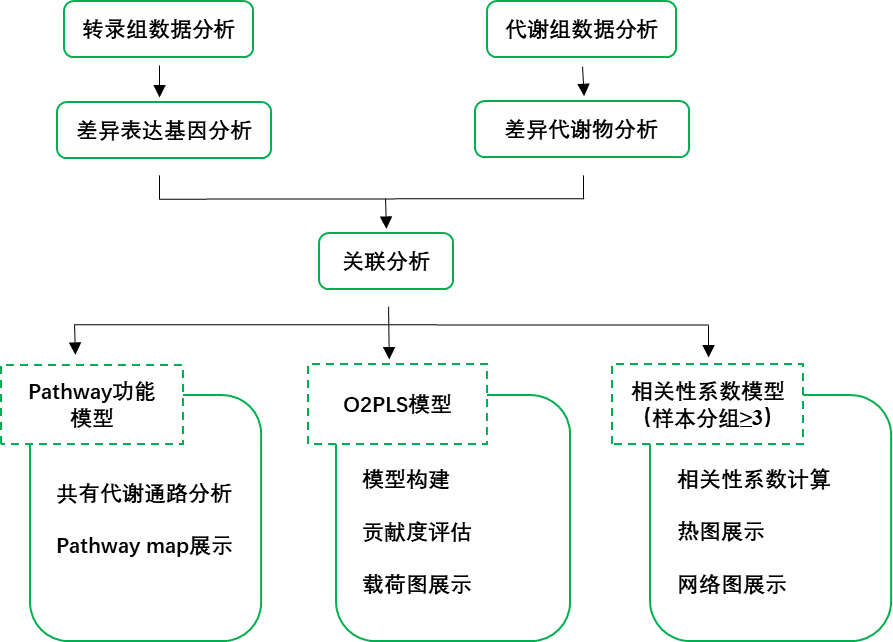
由转录组和代谢组数据获得了样本在基因表达水平和代谢物水平的差异特征。然而生物系统中转录与代谢并不是独立发生的,为揭示基因表达与代谢物之间的调控影响机制,可基于“参与同一生物过程的基因或代谢物具有相同或相似的变化规律”进行二者的关联分析。
3 Pathway功能模型
3.1 共有代谢通路分析
通过组间差异分析,由转录组数据获得了差异表达的基因,由代谢组数据获得了差异表达的代谢物,并进行了各自组学的KEGG富集分析。在关联分析中,将针对基因与代谢物进行共有KEGG pathway的分析[1][2][3]。包含三种类型:
1)组间差异基因与组间差异代谢物共有pathway的分析;
2)由于组间差异代谢物的种类可能很少,或者差异代谢物中没有目标代谢物,故进行组间差异基因与所有代谢物共有pathway的分析;
3)作为其他个性化分析筛选的基础,进行所有基因与所有代谢物共有pathway的分析。
| 关联类型 | 基因数据来源 | 代谢物数据来源 | 共有pathway | Pathway 注释表 |
|---|---|---|---|---|
| 差异基因与差异代谢物 | LN-vs-LL | LN-vs-LL | LN-vs-LL.htm | LN-vs-LL.path.xls |
| TN-vs-TL | TN-vs-TL | TN-vs-TL.htm | TN-vs-TL.path.xls | |
| LL-vs-TL | LL-vs-TL | LL-vs-TL.htm | LL-vs-TL.path.xls | |
| LN-vs-TN | LN-vs-TN | LN-vs-TN.htm | LN-vs-TN.path.xls | |
| 差异基因与所有代谢物 | LN-vs-LL | all | LN-vs-LL_all.htm | LN-vs-LL_all.path.xls |
| TN-vs-TL | all | TN-vs-TL_all.htm | TN-vs-TL_all.path.xls | |
| LL-vs-TL | all | LL-vs-TL_all.htm | LL-vs-TL_all.path.xls | |
| LN-vs-TN | all | LN-vs-TN_all.htm | LN-vs-TN_all.path.xls | |
| 所有基因与所有代谢物 | all | all | all.htm | all.path.xls |
3.2 Pathway map
点击共有pathway分析页面的KEGG描述,可链接基因与代谢物相关联的pathway map,示例如下:
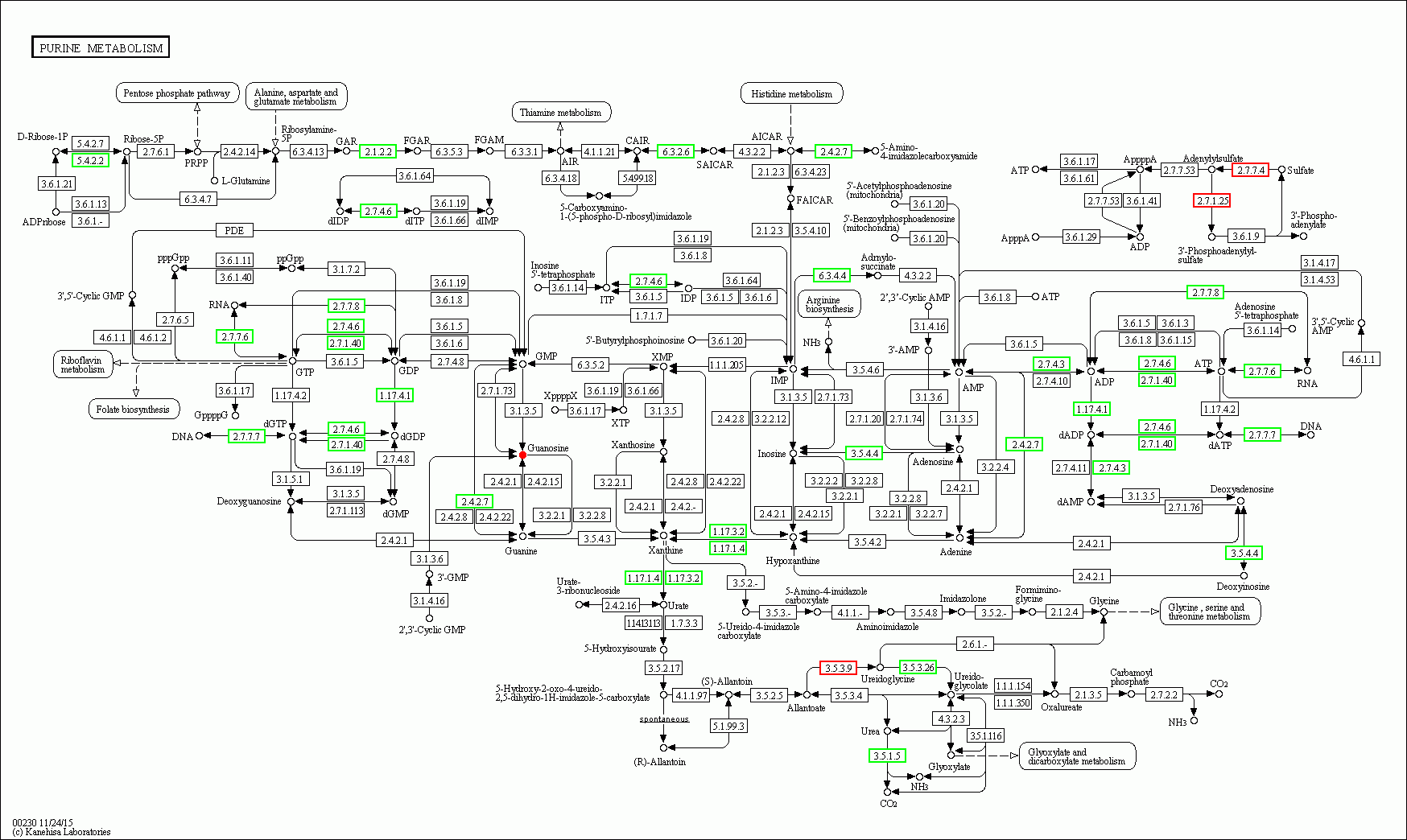 |
Fig 3-2-1 差异代谢物与差异基因关联示例 |
4 O2PLS模型分析
基于所有转录组和所有代谢组数据,我们利用OmicsPLS包开展O2PLS分析[4]。O2PLS模型适用于两个组学数据的关联分析,可在两个数据矩阵中进行双向建模和预测。O2PLS模型通过计算,将每个组学的数据都分解为三个部分,即关联部分(joint part,即与另一组学关联程度很大的部分)、正交部分(Systematic/Orthogonal part,即只对本组学数据有影响、而对另一组学数据没有影响的部分)、噪音部分(noise part,即对两组学数据都没有影响的部分),如下图所示。各部分对总变异的解释程度以R2表示,值越高表示模型的解释能力越好。模型的过拟合和不足都会降低R2。
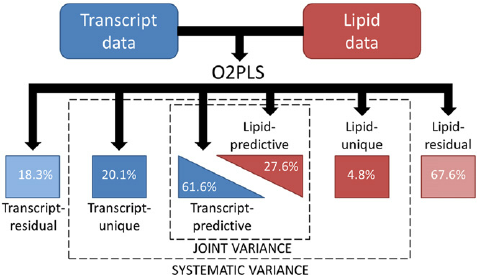 |
| Fig 4-0-1 o2pls模型示例 |
OmicPLS包进行O2PLS模型分析的步骤如下:
1)对两个独立数据集(X,Y,如基因表达量和代谢物丰度数据)分别进行标准化,使总平方和为1,以消除组学数据数量级差异的影响,使统计分析更科学。
2)Cross-validation:确定组分个数。以关联部分组分个数a大于1,基因正交部分组分个数ax和代谢组正交部分组分个数ay大于0开始,进行多次不同数值组合的预建模,计算预测误差。
3)Fitting:最适建模。选取预测误差最小时,对应的组分个数n,nx,ny来构建最适模型。
4)输出各部分贡献度(R2),绘制载荷图。
4.1 模型构建
由于模型的拟合不足或过拟合都会对数据分析造成影响,因此在建立模型之前,我们会对模型进行预评估。模型的关联和正交部分包含一定数量的组分(component,类似PCA分析的主成分PC1、PC2)。我们采用交叉验证法(cross-validation)对每部分所包含的组分个数进行评估与设定。交叉验证法通过多次预建模,选择预测误差(prediction error)最小的模型进行后续分析。
| Model | nx | ny | n | prediction error |
|---|---|---|---|---|
| dif | 0 | 1 | 4 | 0.8159481 |
| LN-vs-LL | 0 | 0 | 4 | 0.8718620 |
| TN-vs-TL | 0 | 0 | 4 | 0.7448241 |
| LL-vs-TL | 0 | 0 | 3 | 0.9848046 |
| LN-vs-TN | 0 | 0 | 4 | 0.5221112 |
4.2 贡献度评估
贡献度是指模型各部分对总变异的解释程度,用R2表示。R2值越高,表示该部分对模型的解释能力越好。
| Model | R2X | R2Y | R2Xcorr | R2Ycorr |
|---|---|---|---|---|
| dif | 0.911 | 0.947 | 0.911 | 0.941 |
| LN-vs-LL | 0.978 | 0.988 | 0.978 | 0.988 |
| TN-vs-TL | 0.978 | 0.987 | 0.978 | 0.987 |
| LL-vs-TL | 0.934 | 0.981 | 0.934 | 0.981 |
| LN-vs-TN | 0.991 | 0.993 | 0.991 | 0.993 |
4.3 载荷图
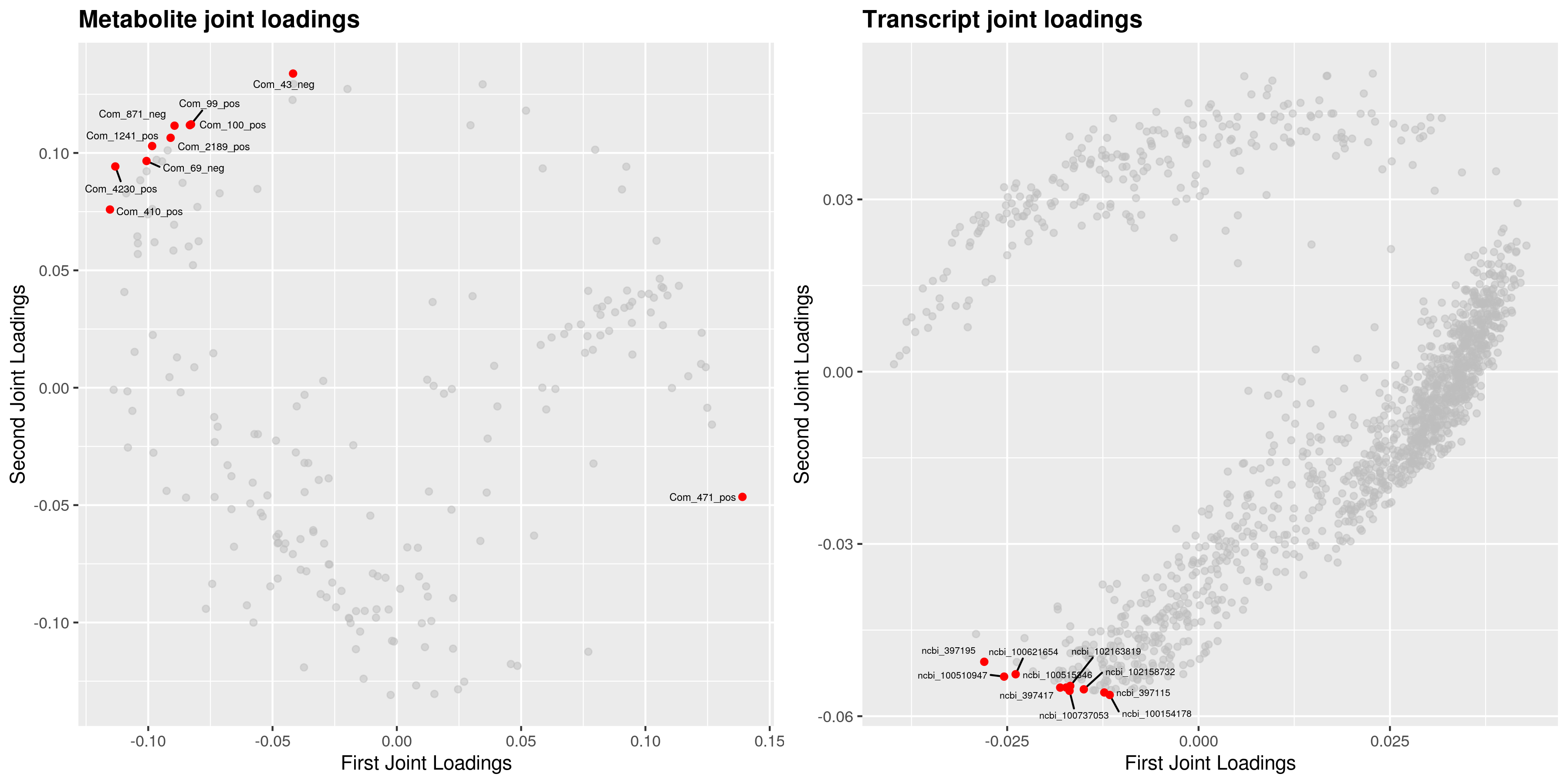
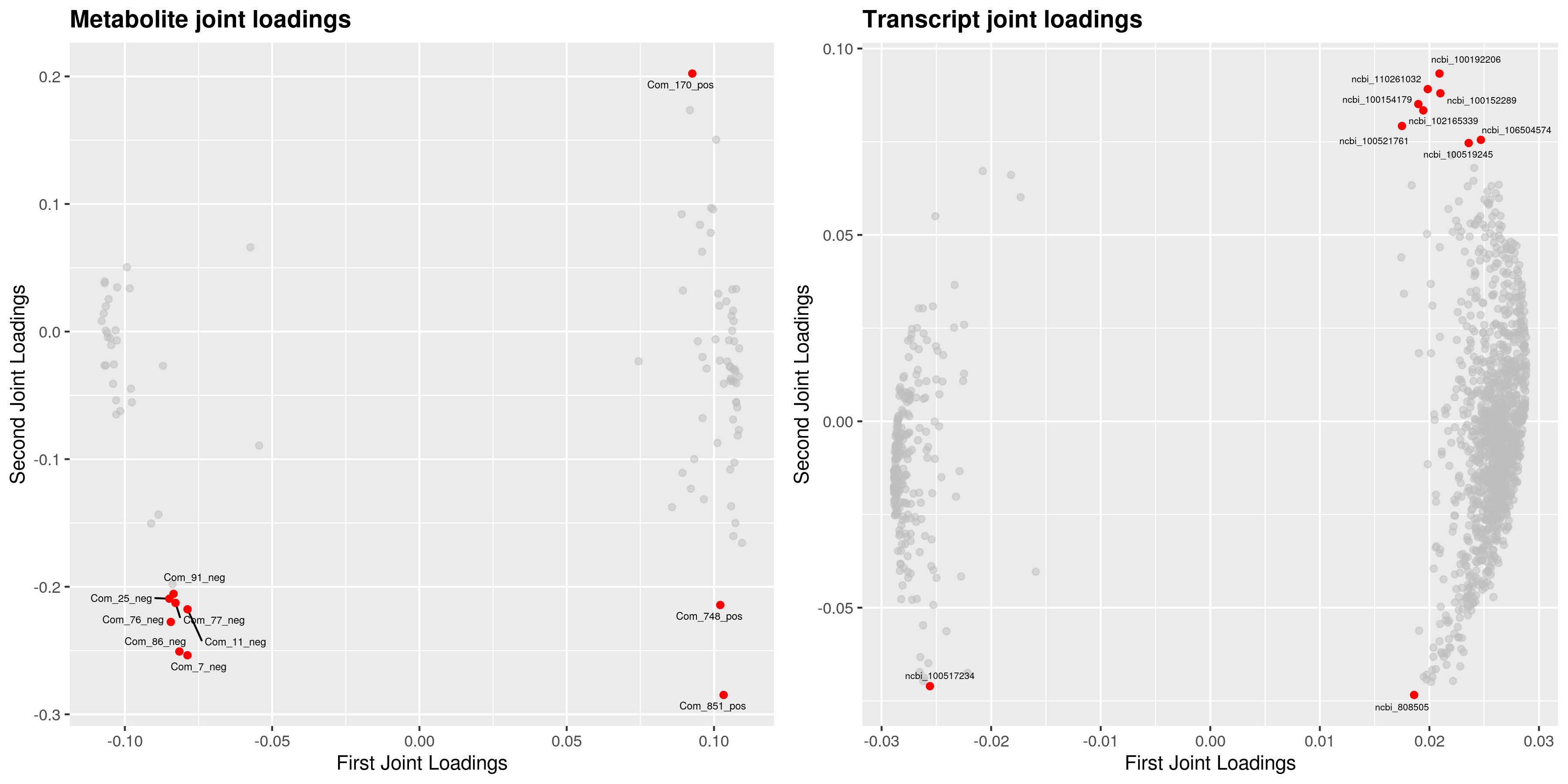
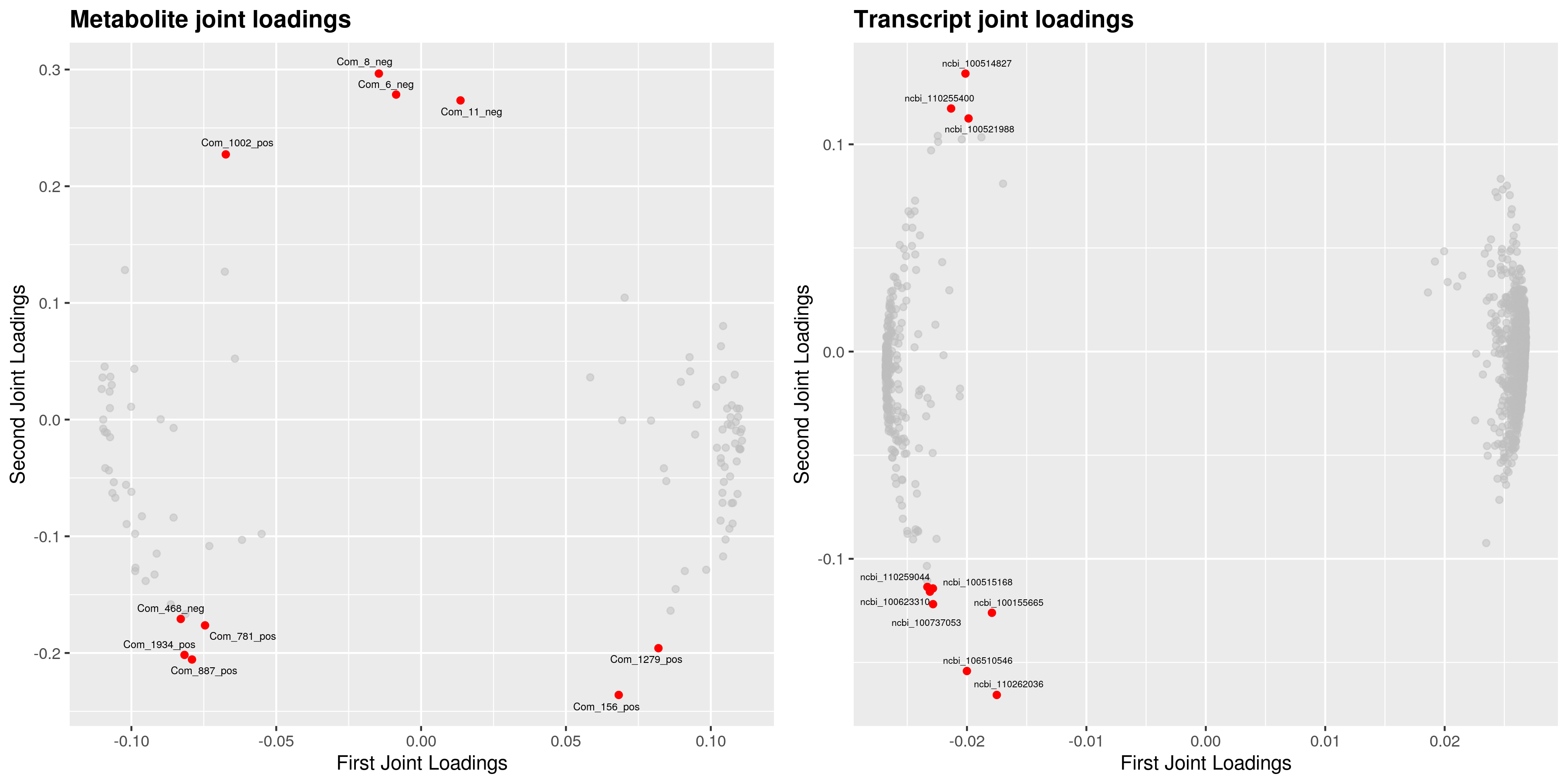
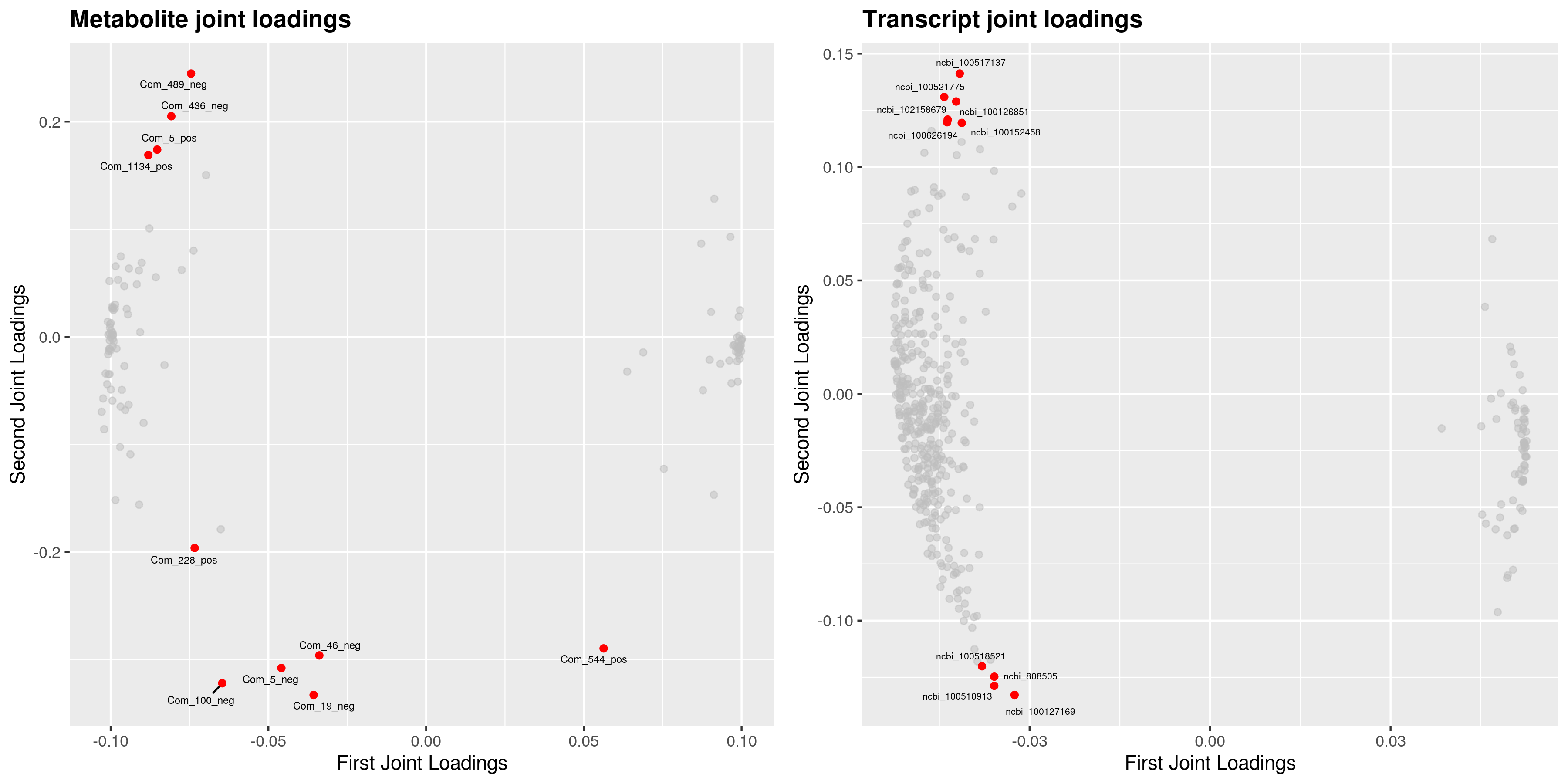
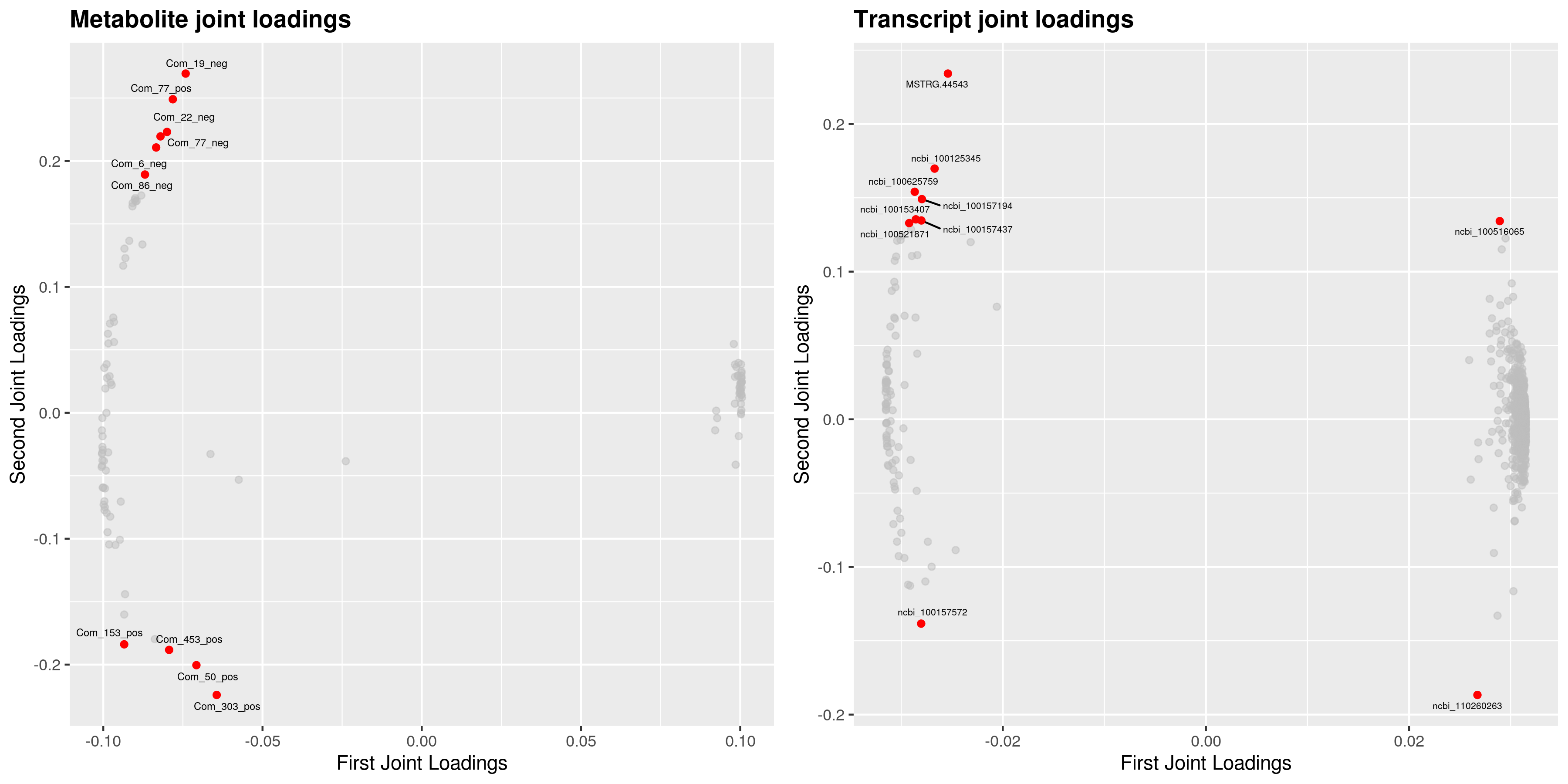
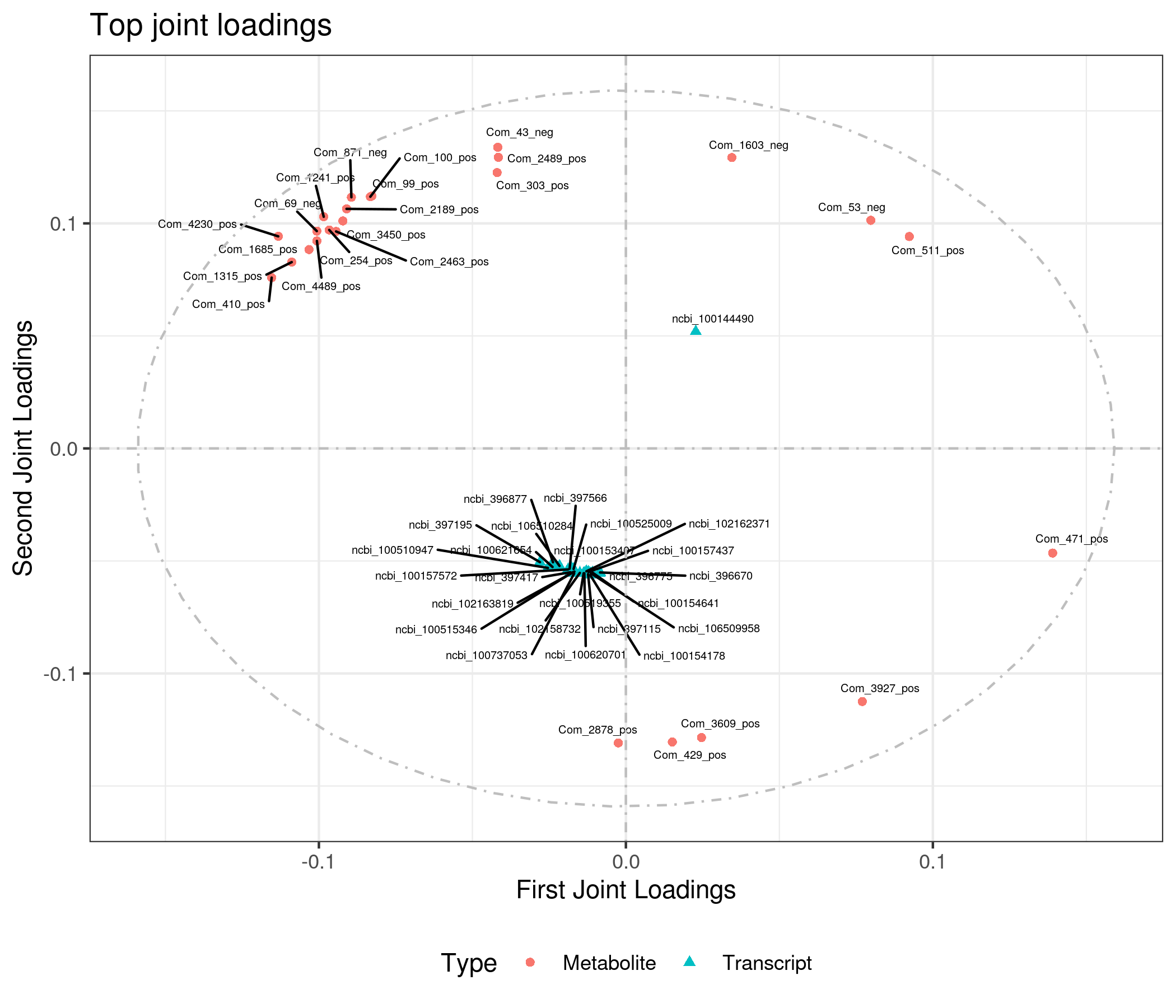
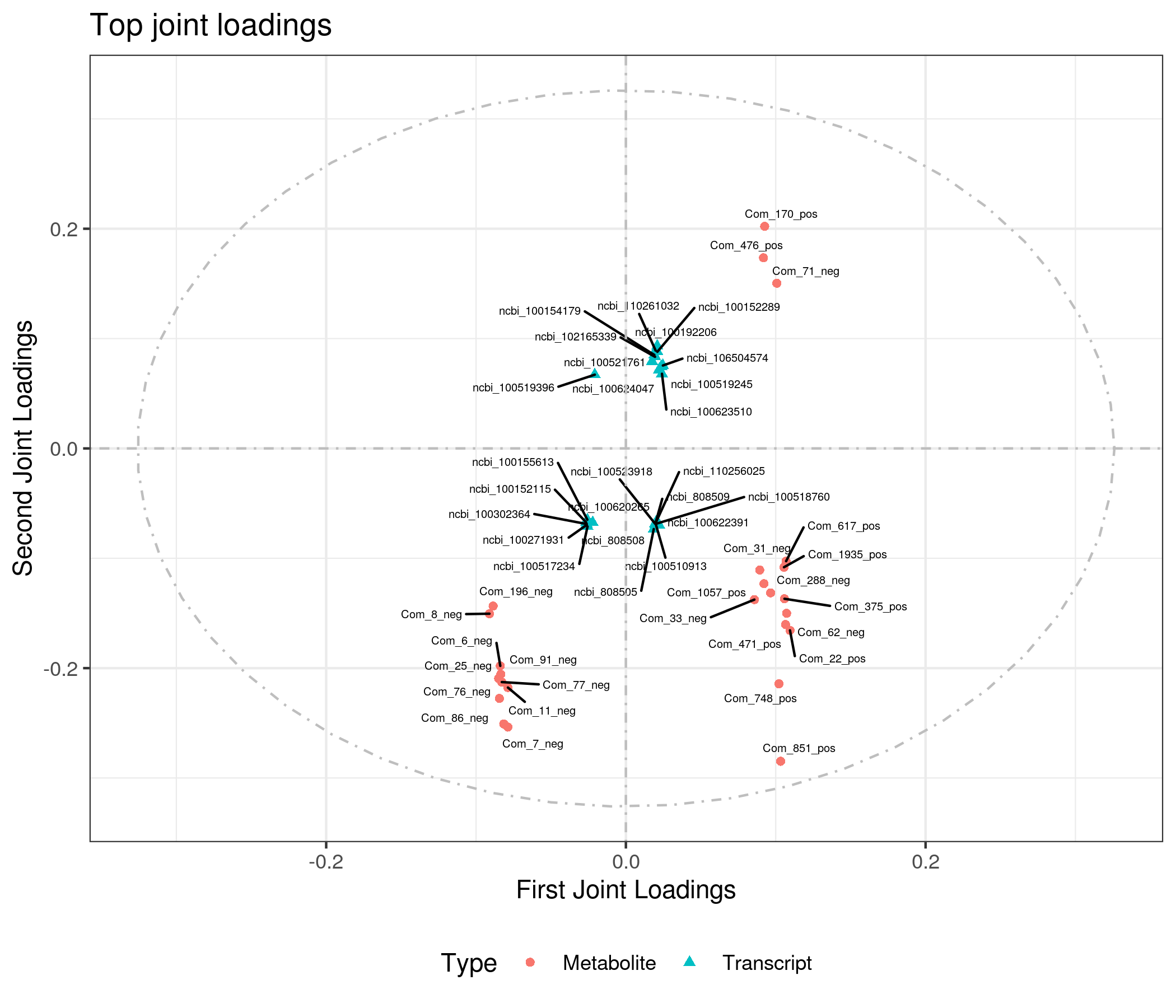
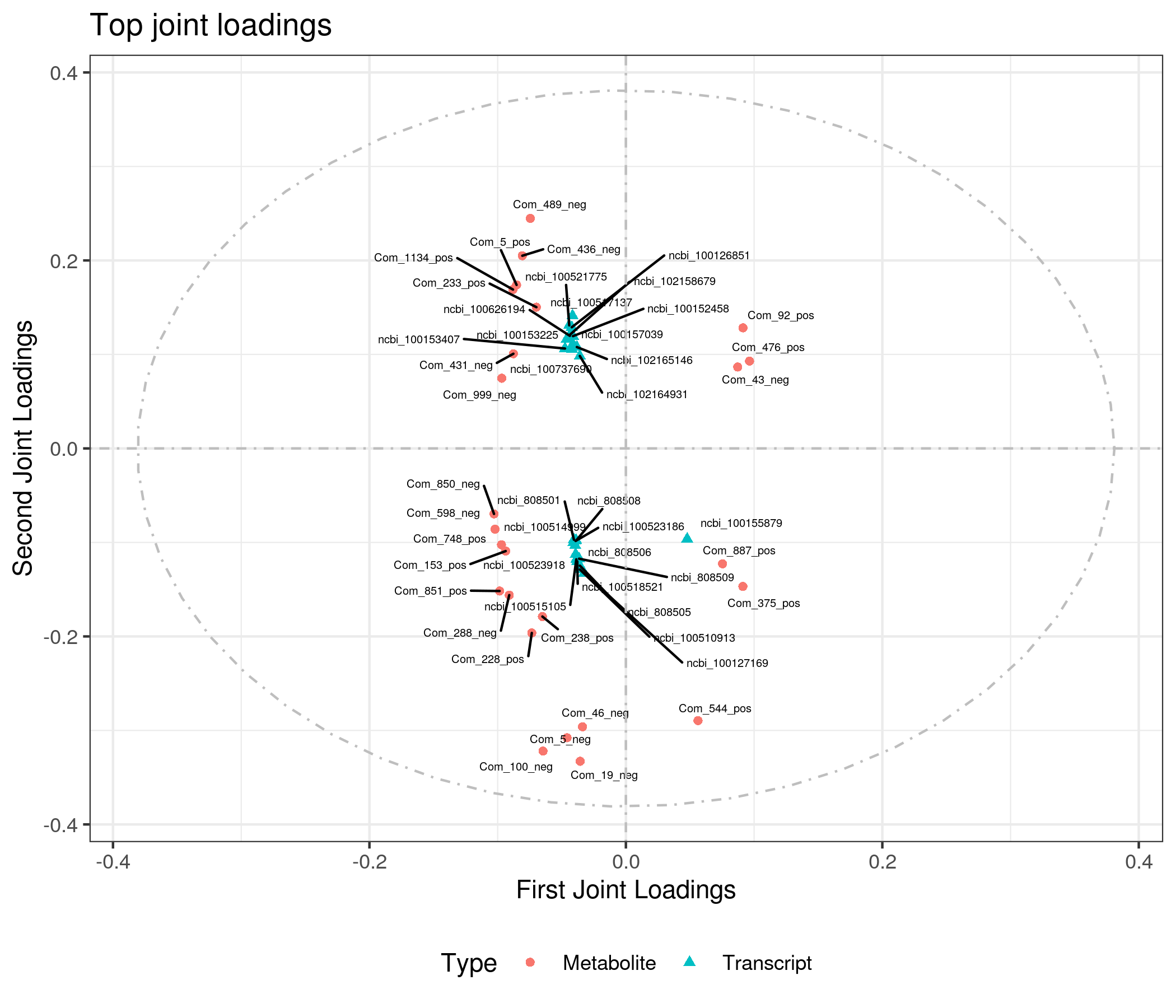
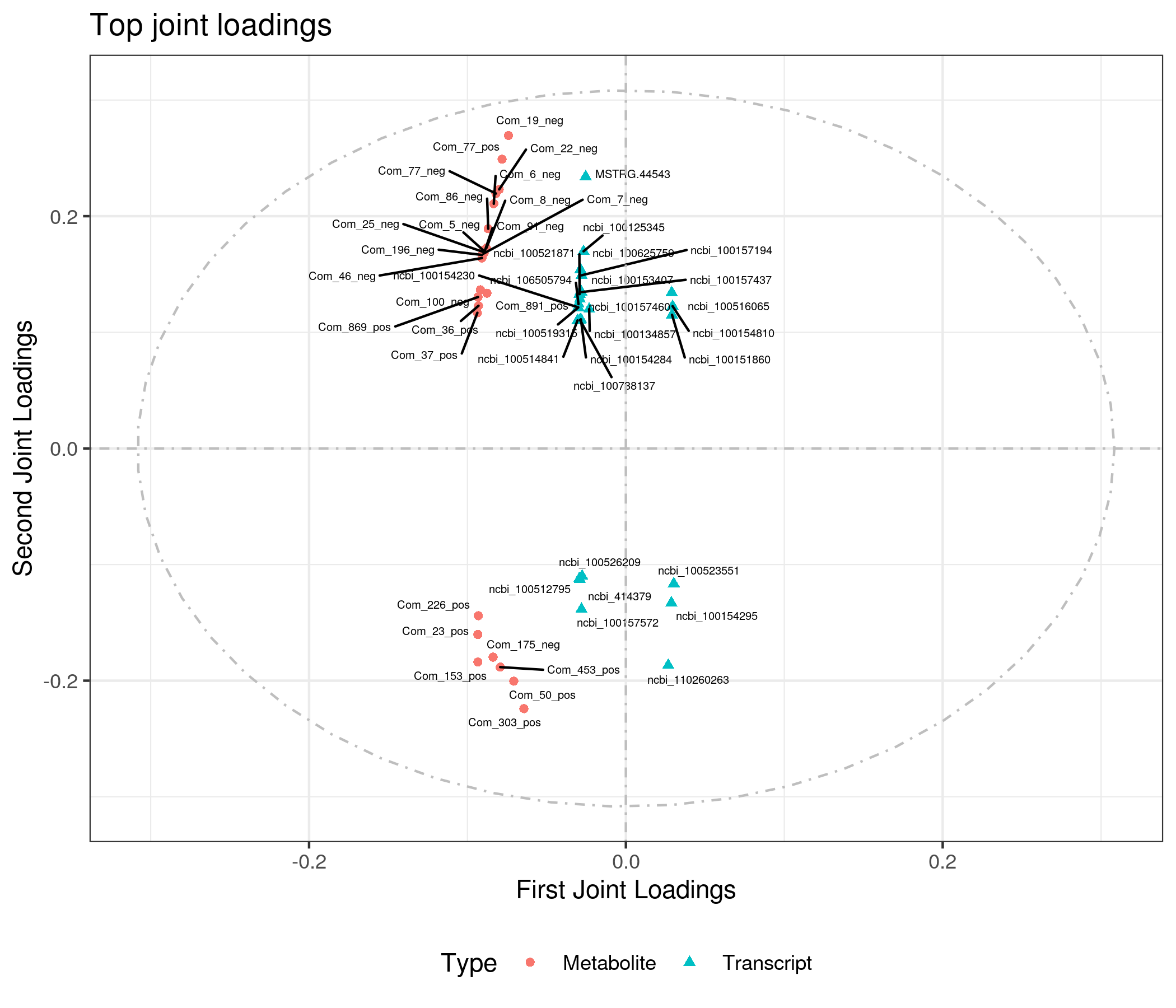
为了查看哪些代谢物和基因相互关联,我们对joint部分的变量分别绘制不同组学的载荷图(loadings plot)。载荷值(loading value)表示变量(代谢物/基因)在各组分的解释能力(即对组间差异的贡献度),载荷值的正负表示与另一组学正关联或负相关;载荷值的绝对值越大,表示关联越强。
4.4 组间元素关联分析
根据元素loading值结果,我们筛选出前两个维度loading值平方和前25的基因和代谢物进行整合绘制loading图,以展示关联程度最大的基因和代谢物。
差异代谢物与差异基因(dif)O2PLS分析loading值结果如下:
5 相关性系数模型
5.1 Pearson系数计算
皮尔逊相关系数(Pearson correlation coefficient)可用来度量两个变量之间的相互关系,代表了两个变量共变性的强弱,取值范围为[-1,+1]。计算基因表达量与代谢物丰度的pearson系数,以评估基因与代谢物的相关性[6][7][8]。
包含两种类型:
1)所有差异基因(各比较组间差异基因的并集)表达量与所有差异代谢物(各比较组间差异代谢物的并集)丰度的pearson系数;
2)所有差异基因(各比较组间差异基因的并集)表达量与所有代谢物丰度的pearson系数。
具体分析结果如下:
| 关联类型 | 基因数据来源 | 代谢物数据来源 | 系数列表 | 热图 |
|---|---|---|---|---|
| 差异基因与 差异代谢物 | 转录组差异集合 | 代谢组差异集合 | dif.pearson.xls | dif.heatmap.corMatrix.png |
| 差异基因与 所有代谢物 | 转录组差异集合 | 所有代谢物 | dif_all.pearson.xls | dif_all.top250.heatmap.png |
差异基因并集与差异代谢物peason相关性结果如下:
5.2 相关性热图
由于差异基因的数目普遍较多,不利于可视化展示关联特征,故先对所有差异基因进行筛选。取相关系数排名前50的差异基因与差异代谢物的相关性以热图展示:
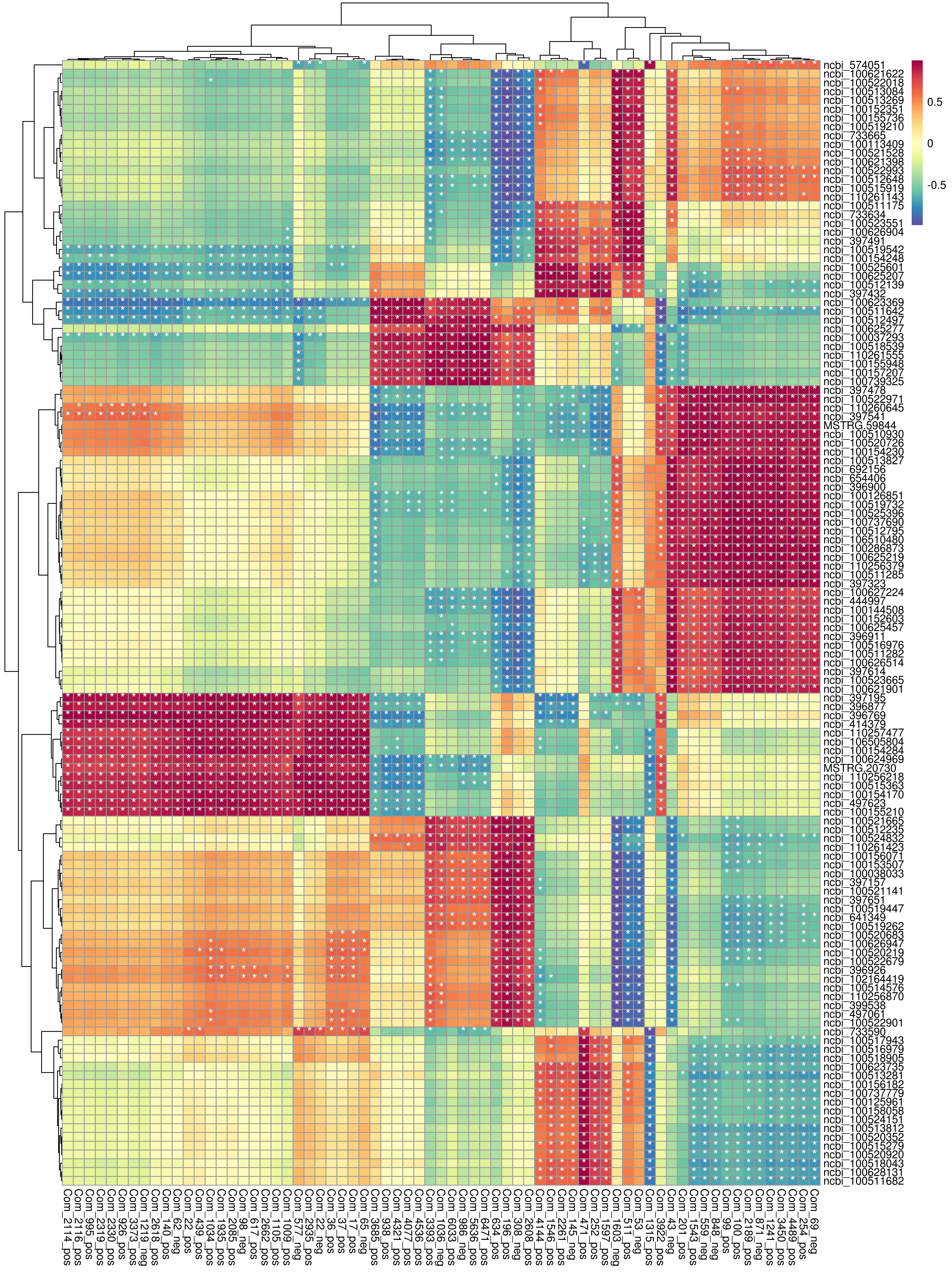 |
| Fig 5-2-1 基因表达量与代谢物丰度相关性热图图 |
5.3 相关性网络图
网络图可以展现处于重要关联位置的基因或代谢物,筛选出相关系数绝对值大于0.5且排名前250的差异基因与差异代谢物数据绘制网络图结果: pearson.sigma_net.html
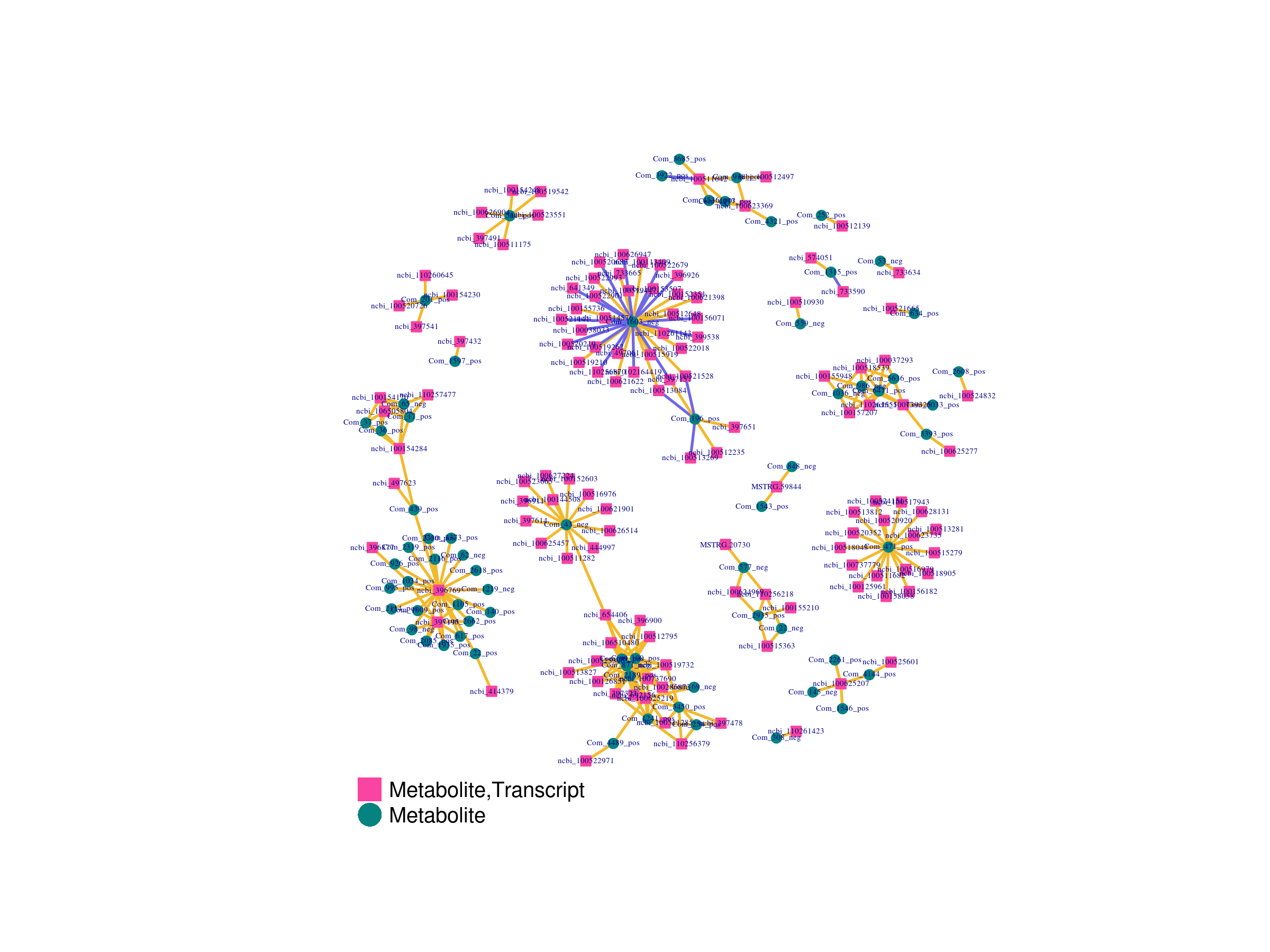 |
Fig 5-3-1 基因表达量与代谢物丰度相关性网络图 |
6 目录结构
├── 01.expression_data [组学丰度数据]
│ ├── groups.txt [分组文件]
│ ├── met.diff.xls [代谢组差异并集丰度]
│ └── rna.diff.xls [转录组差异并集丰度]
├── 02.pathway [pathway模型结果目录]
│ ├── 1.dif_pathway [差异基因与差异代谢物的分析]
│ │ └── Z-vs-ZJ [比较组Z与ZJ的分析]
│ │ ├── Z-vs-ZJ.htm [pathway富集分析]
│ │ ├── Z-vs-ZJ_map [pathway图文件]
│ │ └── Z-vs-ZJ.path.xls [pathway注释]
│ ├── 2.dif_all_pathway [差异基因与所有代谢物的分析]
│ │ └── ZR-vs-ZJR_all [ZR与ZJR差异基因并集与所有代谢物分析]
│ │ ├── ZR-vs-ZJR_all.htm [pathway富集分析]
│ │ ├── ZR-vs-ZJR_all_map [pathway图文件]
│ │ └── ZR-vs-ZJR_all.path.xls [pathway注释]
│ └── 3.all_pathway [所有基因与所有代谢物的分析]
│ ├── all.htm [pathway富集分析]
│ ├── all_map [pathway图文件]
│ └── all.path.xls [pathway注释]
├── 03.o2pls [O2PLS模型结果目录]
│ ├── all.cv.stat.xls [cv分析统计结果]
│ ├── component_proportions.xls [模型组分统计结果]
│ ├── dif [差异基因与差异代谢物o2pls模型]
│ │ ├── dif.component_proportions.xls [模型组分统计]
│ │ ├── dif.cv.stat.xls [cv分析统计]
│ │ ├── o2m_loading.{pdf,png} [模型loading图]
│ │ ├── top25_loading_element.{pdf,png} [loading值前25的关联图]
│ │ ├── dif.Metabiolite_loading.xls [代谢组 loading值表]
│ │ └── dif.Transcript_loading.xls [转录组 loading值表]
│ └── Z-vs-ZJ [Z与ZJ差异基因与差异代谢物o2pls模型]
│ ├── Z-vs-ZJ.component_proportions.xls [cv分析统计]
│ ├── Z-vs-ZJ.cv.stat.xls [模型组分统计]
│ ├── o2m_loading.{pdf,png} [模型loading图]
│ ├── top25_loading_element.{pdf,png} [loading值前25的关联图]
│ ├── Z-vs-ZJ.Metabiolite_loading.xls [代谢组 loading值表]
│ └── Z-vs-ZJ.Transcript_loading.xls [转录组 loading值表]
├── 04.correlation [peason相关系数模型]
│ ├── 1.dif_pearson [差异基因与差异代谢物相关性结果]
│ │ ├── dif.pearson.xls [peason相关性结果]
│ │ ├── dif.top250.heatmap.{pdf,png} [相关性前250基因与代谢物热图]
│ │ ├── dif.top250.corMatrix.xls [相关性前250基因与代谢物矩阵]
│ │ ├── top250.{Node,Edge}4cytoscape.tsv [相关性前250网络图绘图数据]
│ │ ├── pearson.sigma_net.html [相关系数网路图]
│ │ └── pearson.sigma_net.png [相关系数网路图]
│ └── 2.dif_all_pearson [差异基因与所有代谢物相关性结果]
│ ├── dif_all.pearson.xls [peason相关性结果]
│ ├── dif_all.top250.heatmap.{pdf,png} [相关性前250基因与代谢物热图]
│ ├── dif_all.top250.corMatrix.xls [相关性前250基因与代谢物矩阵]
│ ├── top250.{Node,Edge}4cytoscape.tsv [相关性前250网络图绘图数据]
│ ├── pearson.sigma_net.html [相关系数网路图]
│ └── pearson.sigma_net.png [相关系数网路图]
├── index.html [html版结题报告索引]
└── src [结题报告配置文件夹]
├── content.html [结题报告主体html文件]
├── css [css配置文件夹]
├── doc [说明文档配置文件夹]
├── image [图片配置文件夹]
└── js [js配置文件夹]
7 附录
7.1 参考文献
[1] Cho K, Cho K, Sohn H et al. 2016. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation[J]. Journal of Experimental Botany, 67(5): 1519-1533.
[2] Zhang X, Zhou Q, Zou W et al. 2017. Molecular Mechanisms of Developmental Toxicity Induced by Graphene Oxide at Predicted Environmental Concentrations[J]. Environ Sci Technol, 51(14): 7861-7871.
[3] Li Q, Guo S, Jiang X et al. 2016. Mice carrying a human GLUD2 gene recapitulate aspects of human transcriptome and metabolome development[J]. Proceedings of the National Academy of Sciences, 113(19): 5358-5363.
[4] El Bouhaddani, Said, et al. "Evaluation of O2PLS in Omics data integration." BMC bioinformatics. Vol. 17. No. 2. BioMed Central, 2016.
[5] Szymanski, Jedrzej, et al. "Linking gene expression and membrane lipid composition of Arabidopsis." The Plant Cell26.3 (2014): 915-928.
[6] Hamanishi E T, Barchet G L, Dauwe R et al. 2015. Poplar trees reconfigure the transcriptome and metabolome in response to drought in a genotype- and time-of-day-dependent manner[J]. BMC Genomics, 16(1).
[7] Bartel J, Krumsiek J, Schramm K et al. 2015. The Human Blood Metabolome-Transcriptome Interface[J]. PLOS Genetics, 11(6): e1005274.
[8] Copley T R, Aliferis K A, Kliebenstein D J et al. 2017. An integrated RNAseq-1H NMR metabolomics approach to understand soybean primary metabolism regulation in response to Rhizoctonia foliar blight disease[J]. BMC Plant Biology, 17(1).
7.2 帮助文档
代谢组与转录组关联分析帮助文档:help.html
代谢组与转录组关联分析英文方法:method.pdf
7.3 引用与致谢
如果您的研究课题使用了基迪奥的测序和分析服务,我们期望您在论文发表时,在Method部分或Acknowledgements部分引用或提及基迪奥公司。
以下语句可供参考:
We are grateful to/thank Guangzhou Genedenovo Biotechnology Co., Ltd for assisting in sequencing and/or bioinformatics analysis.