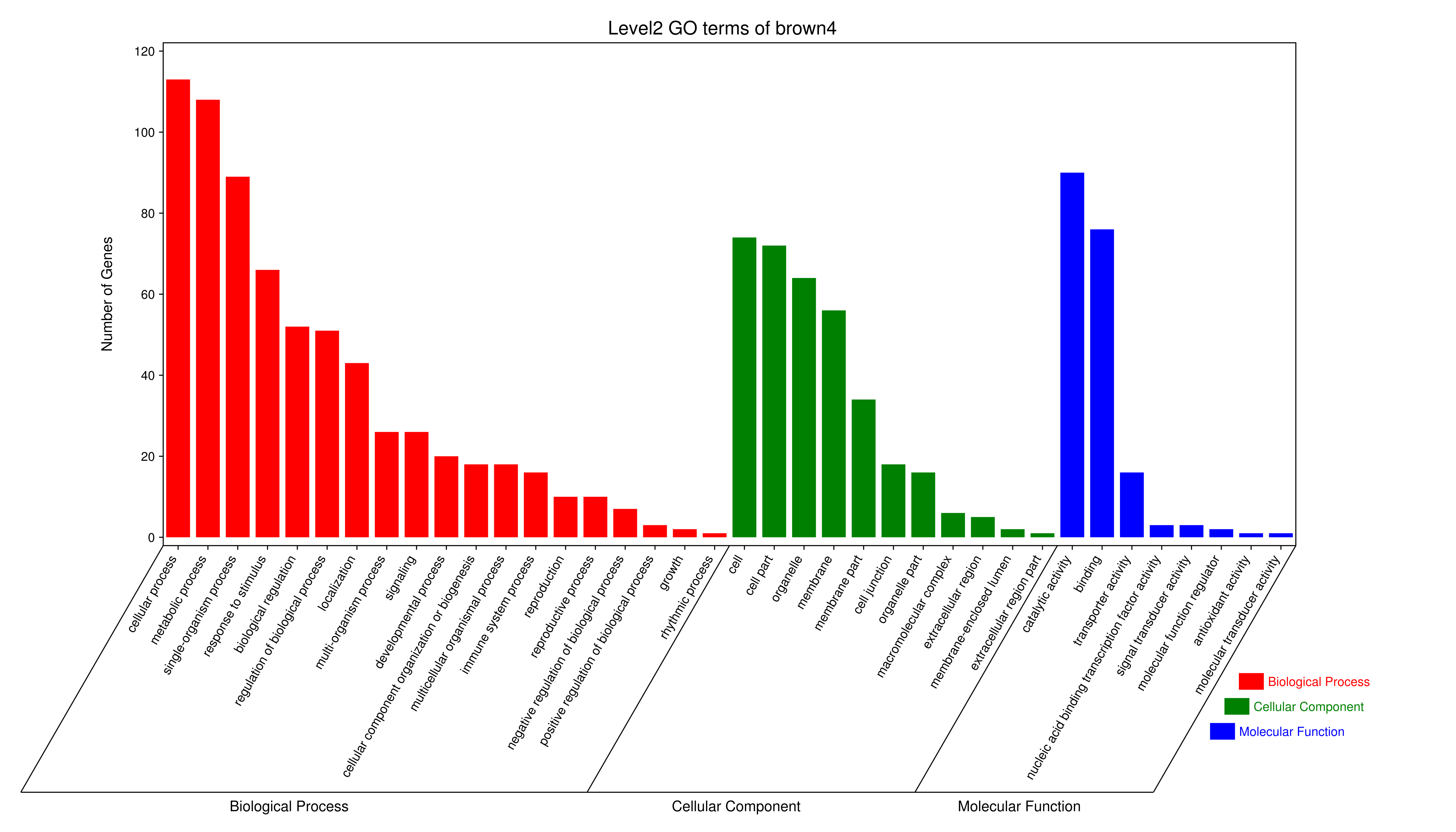
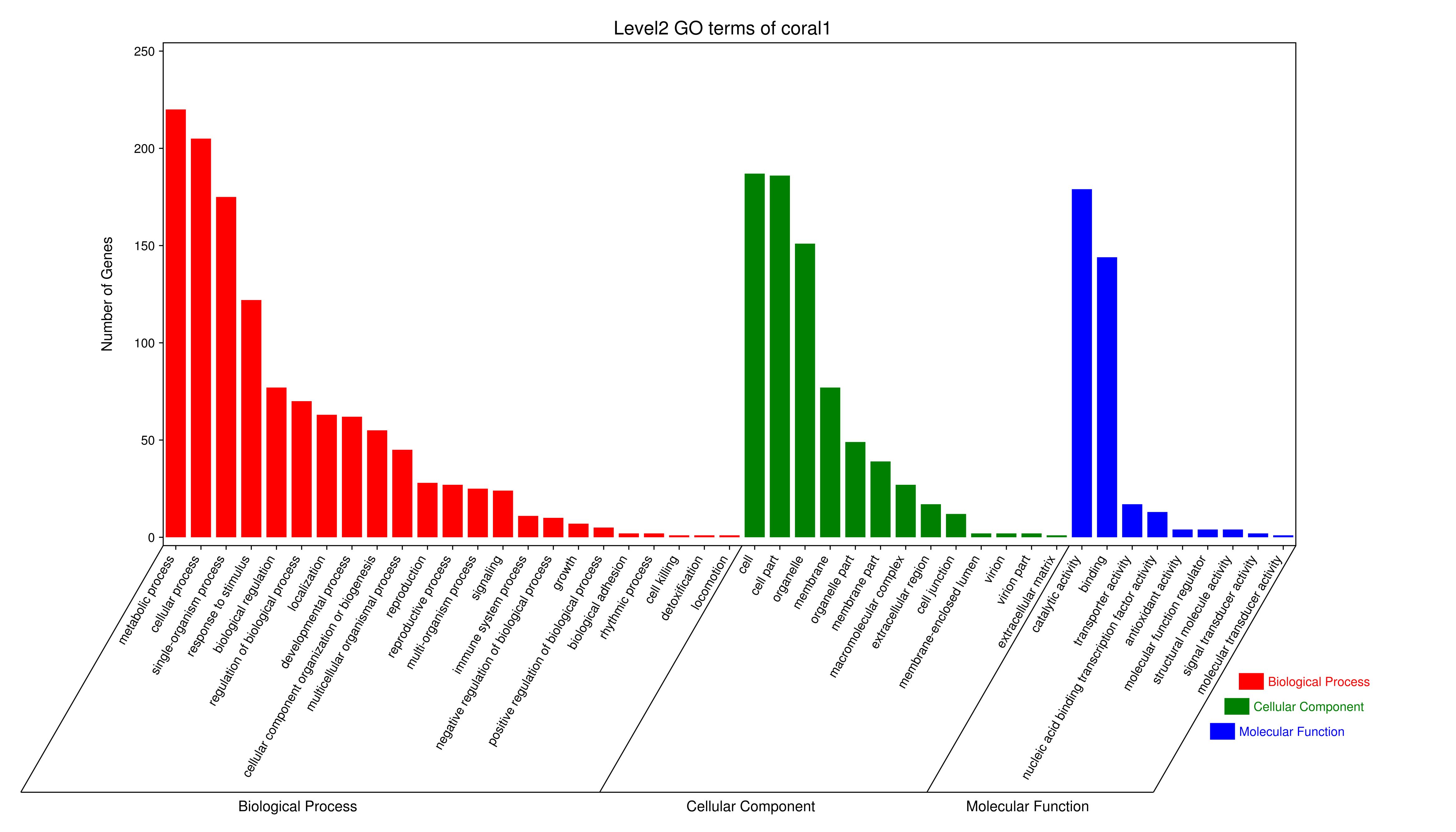
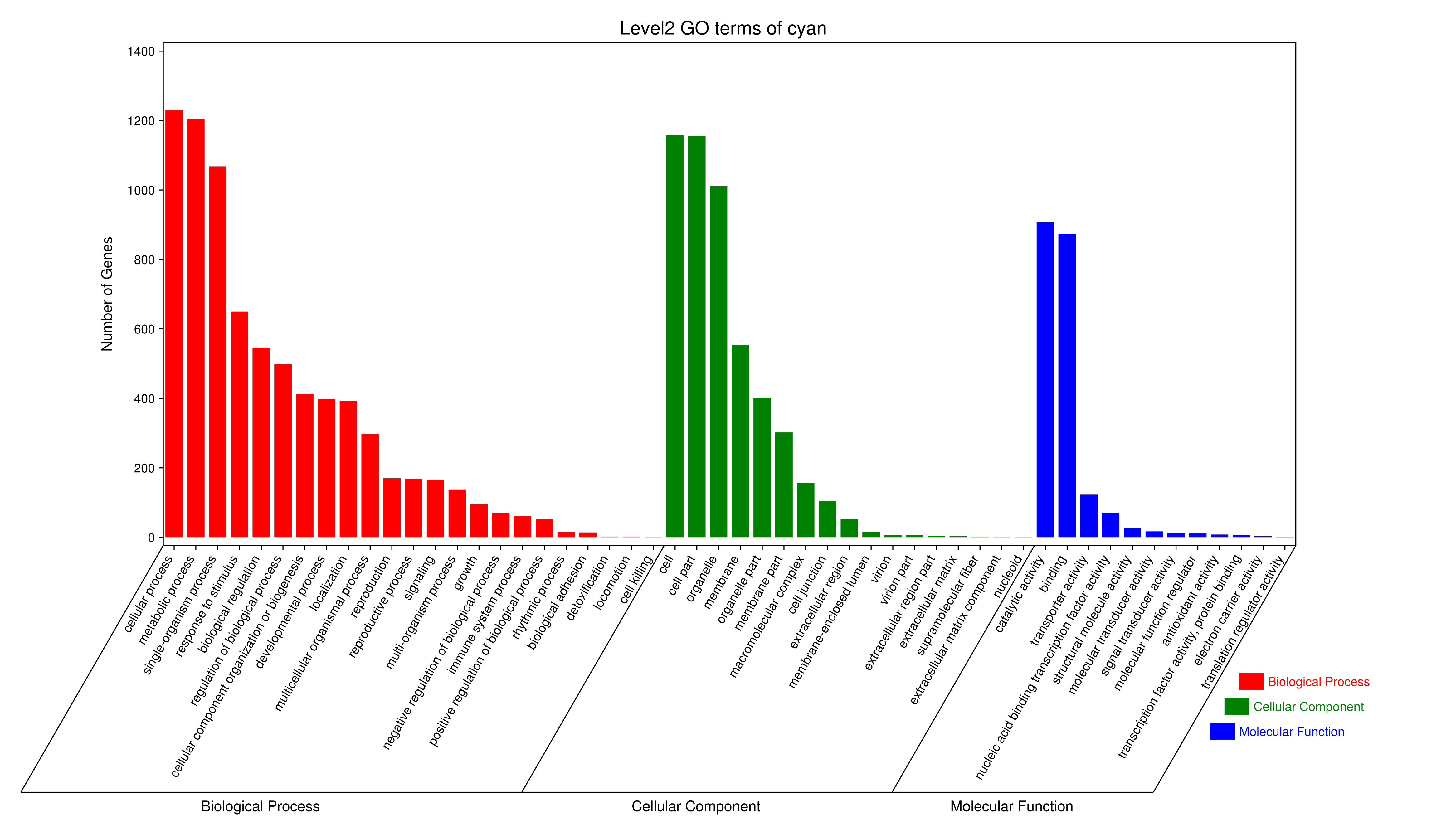
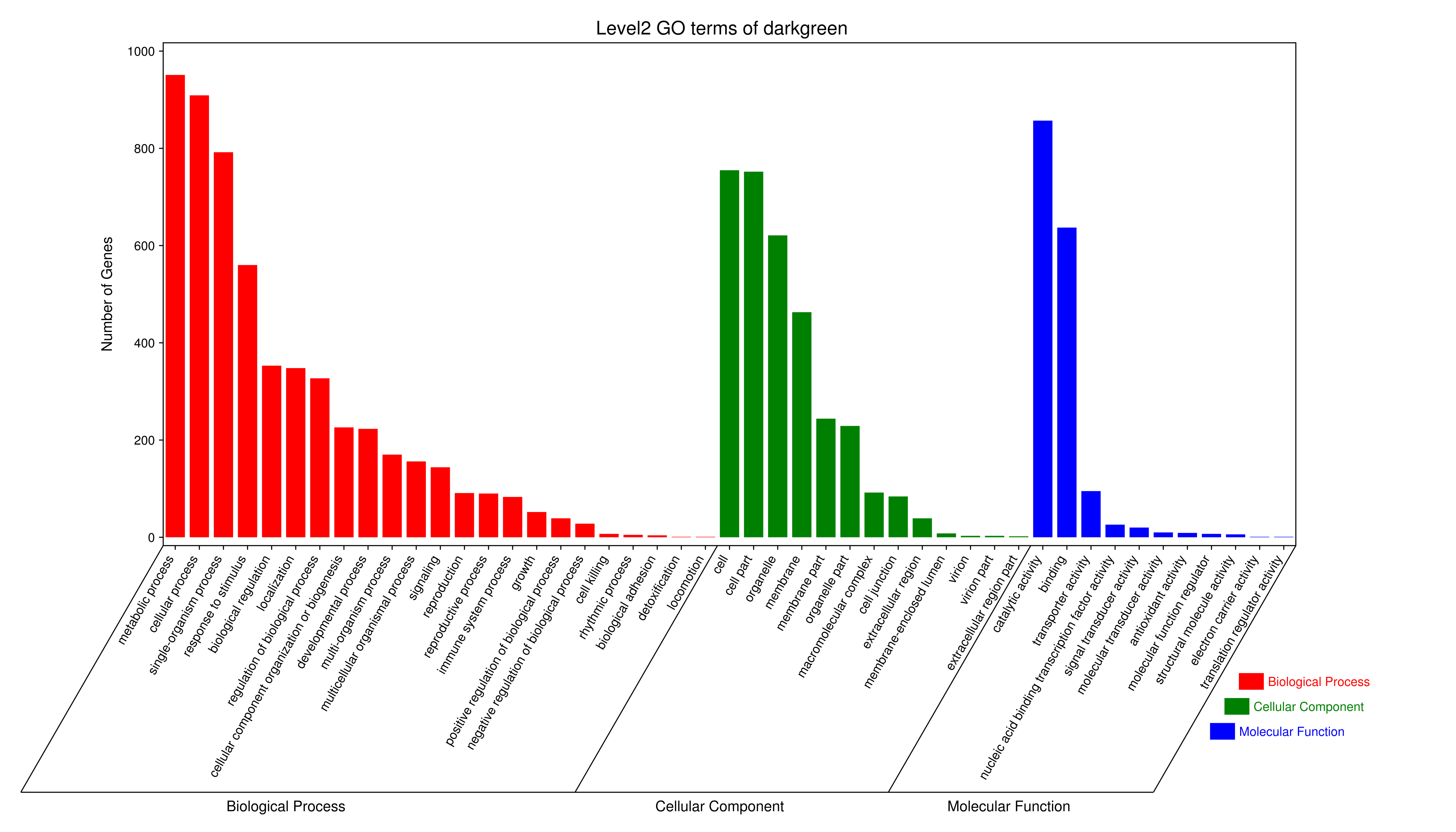
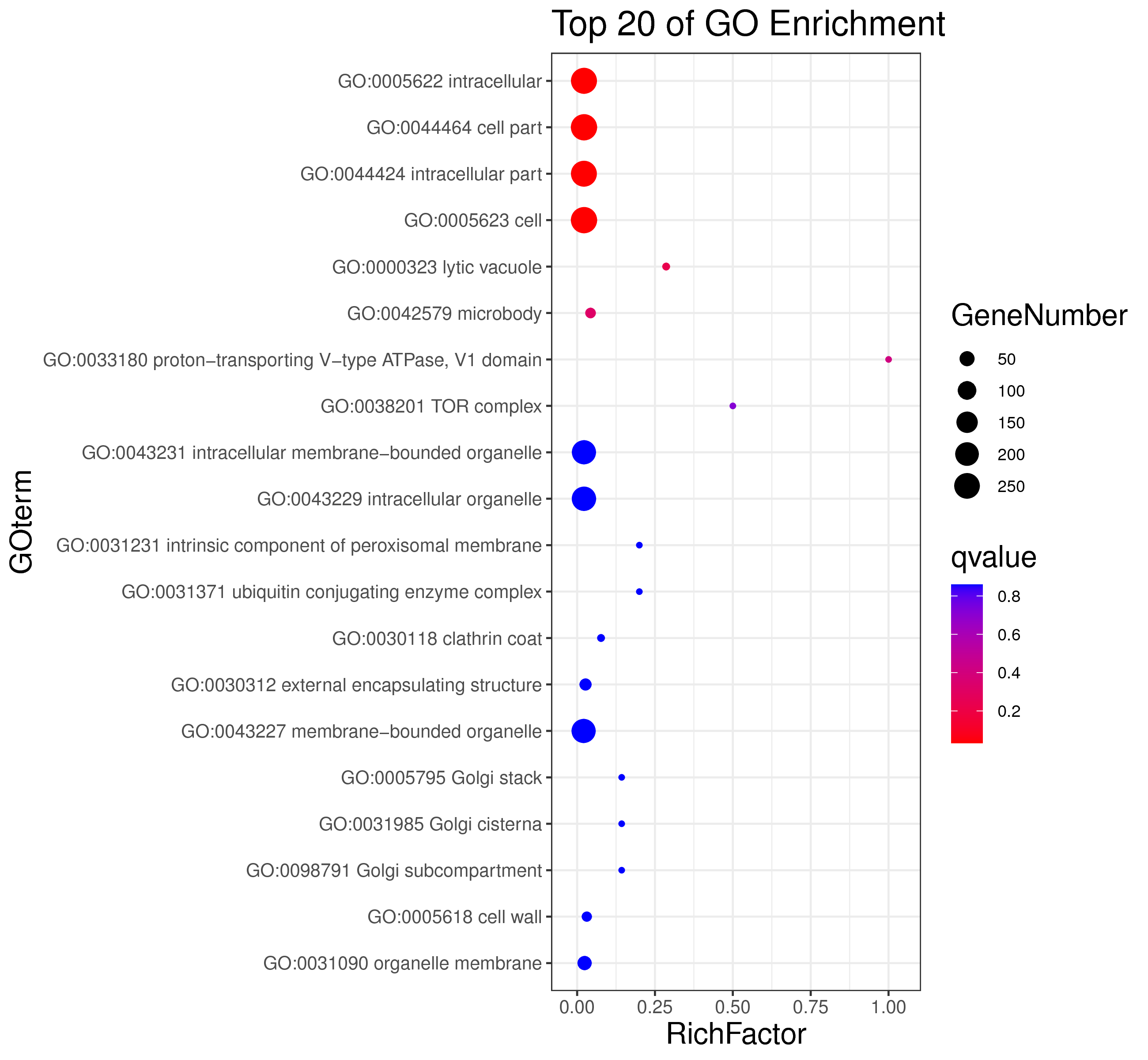
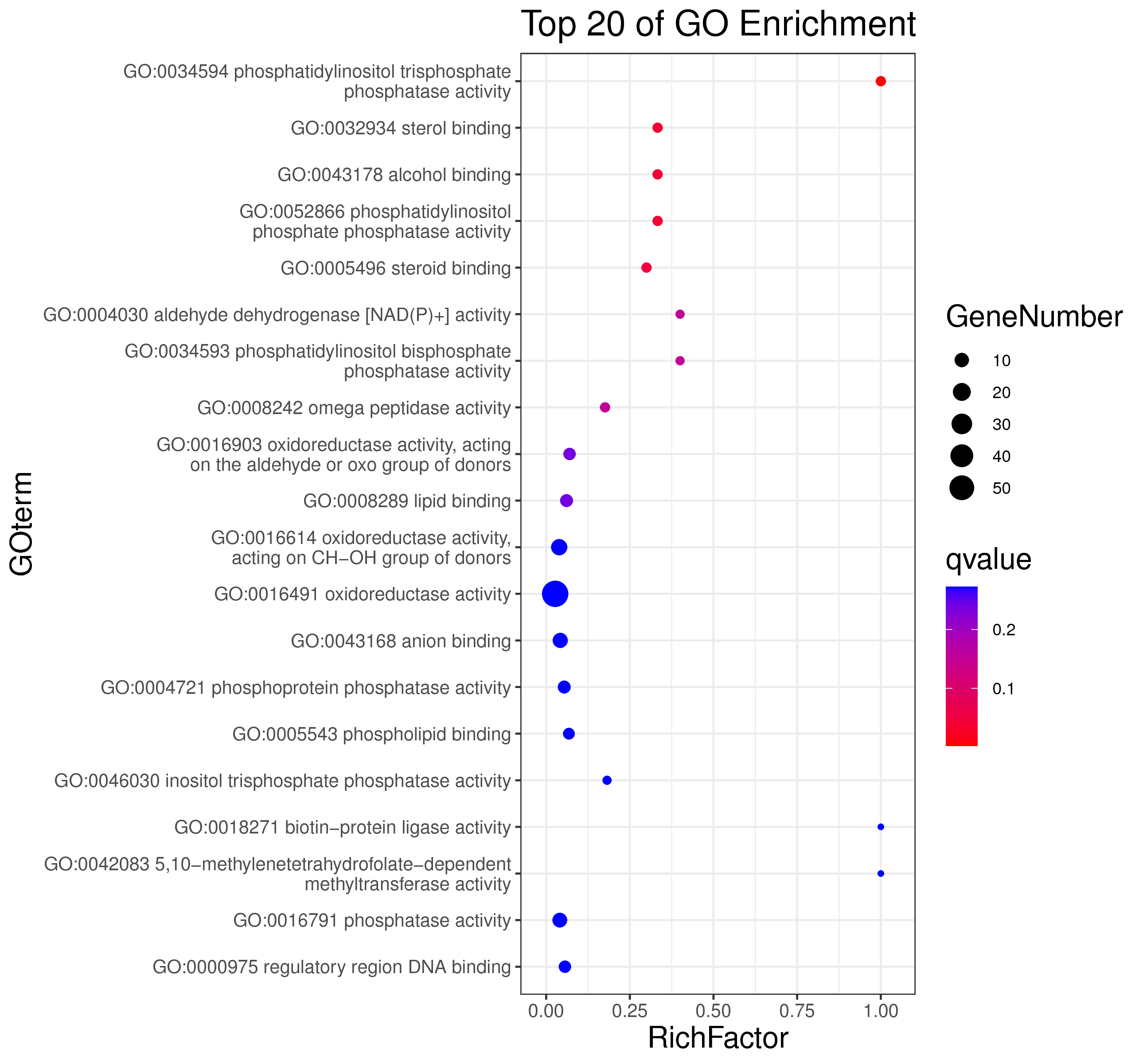
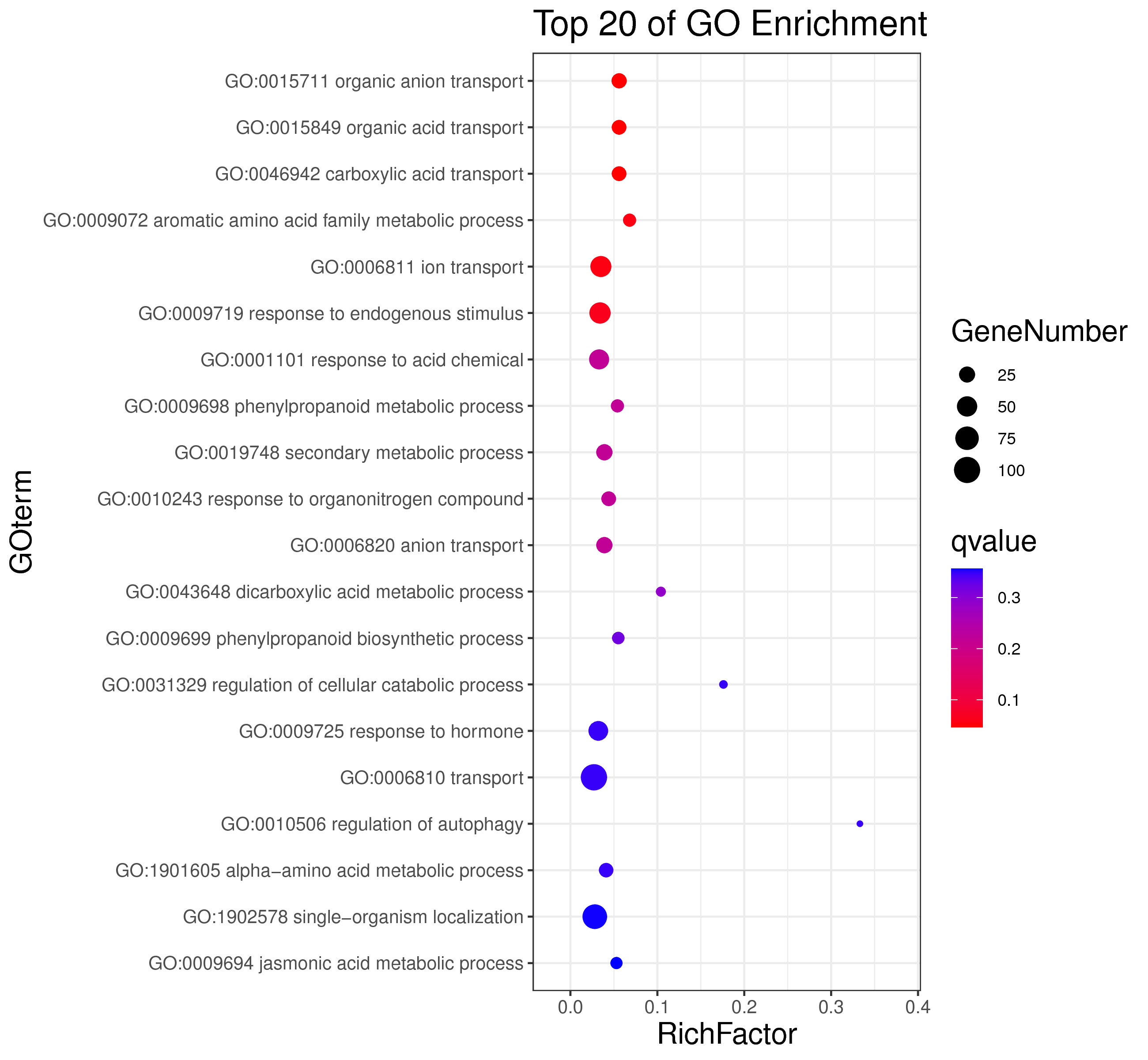
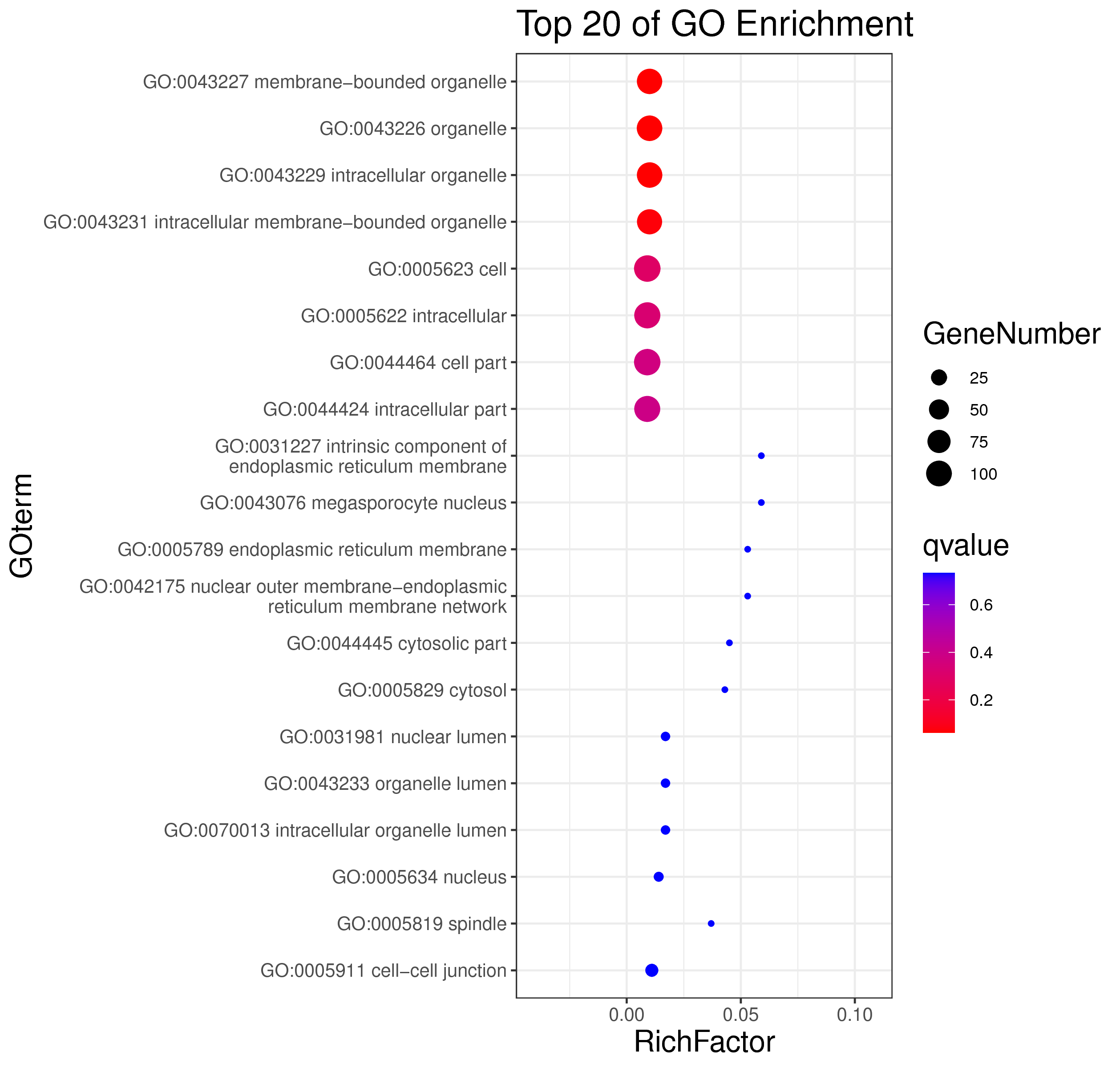
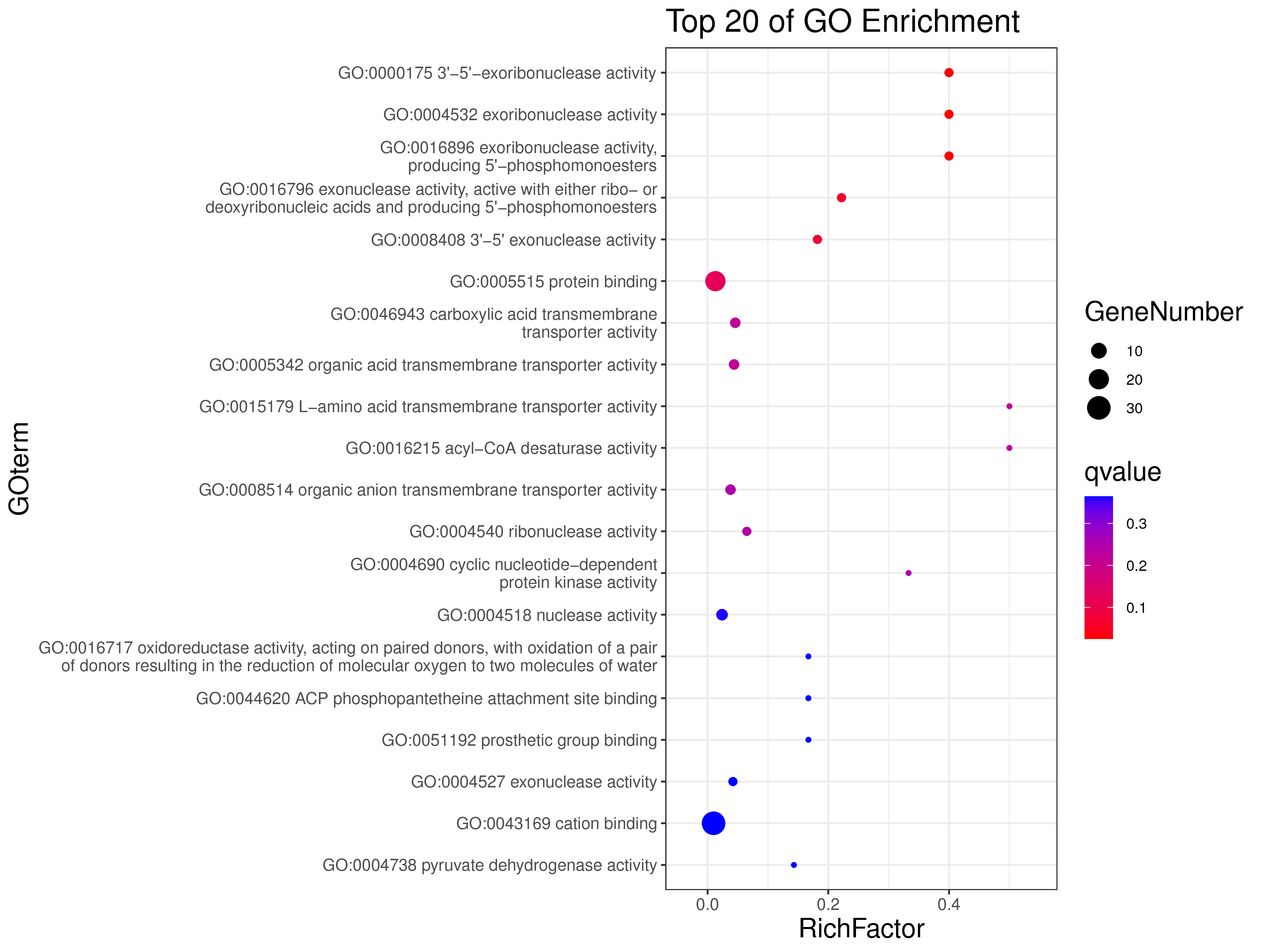
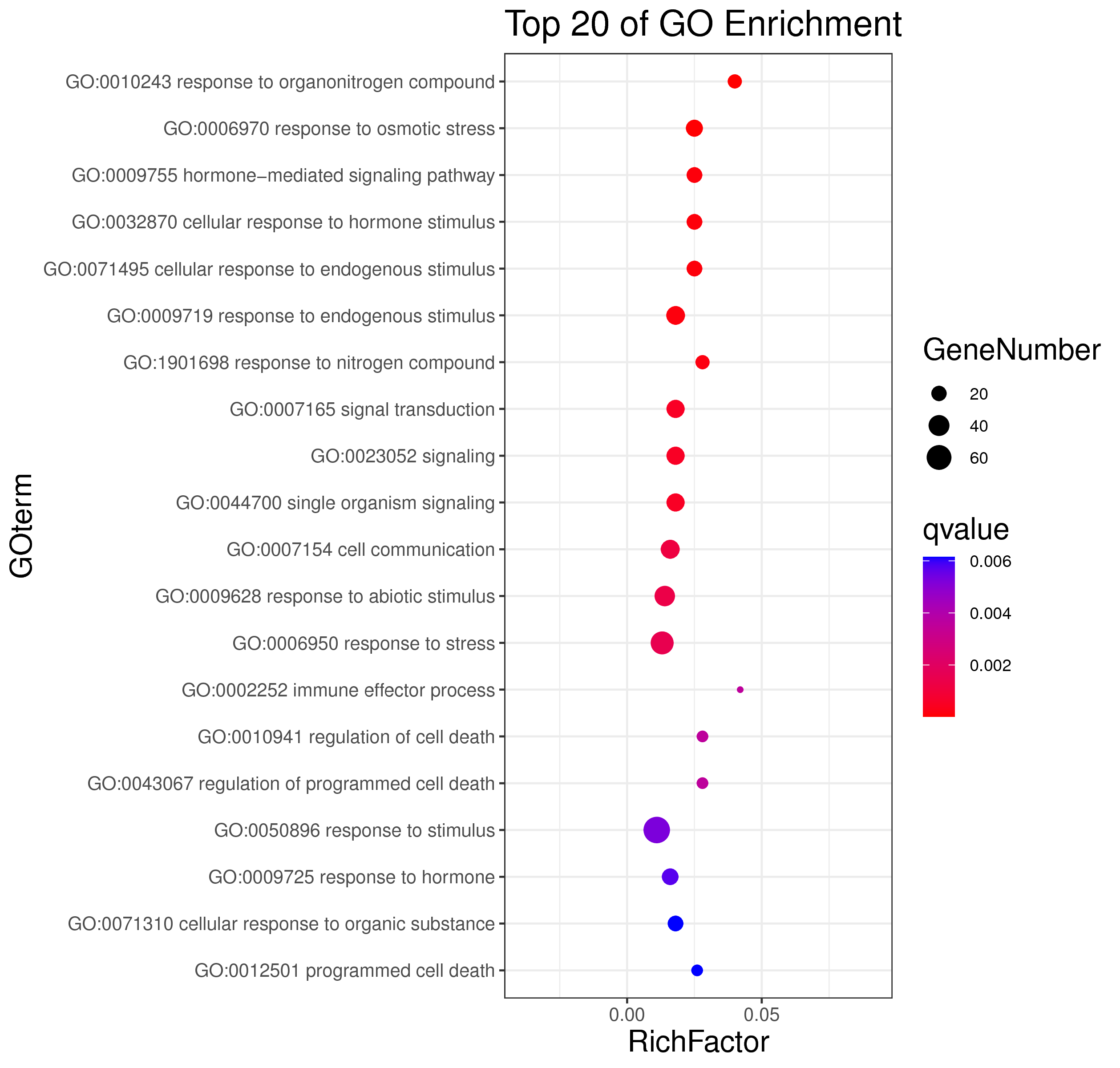
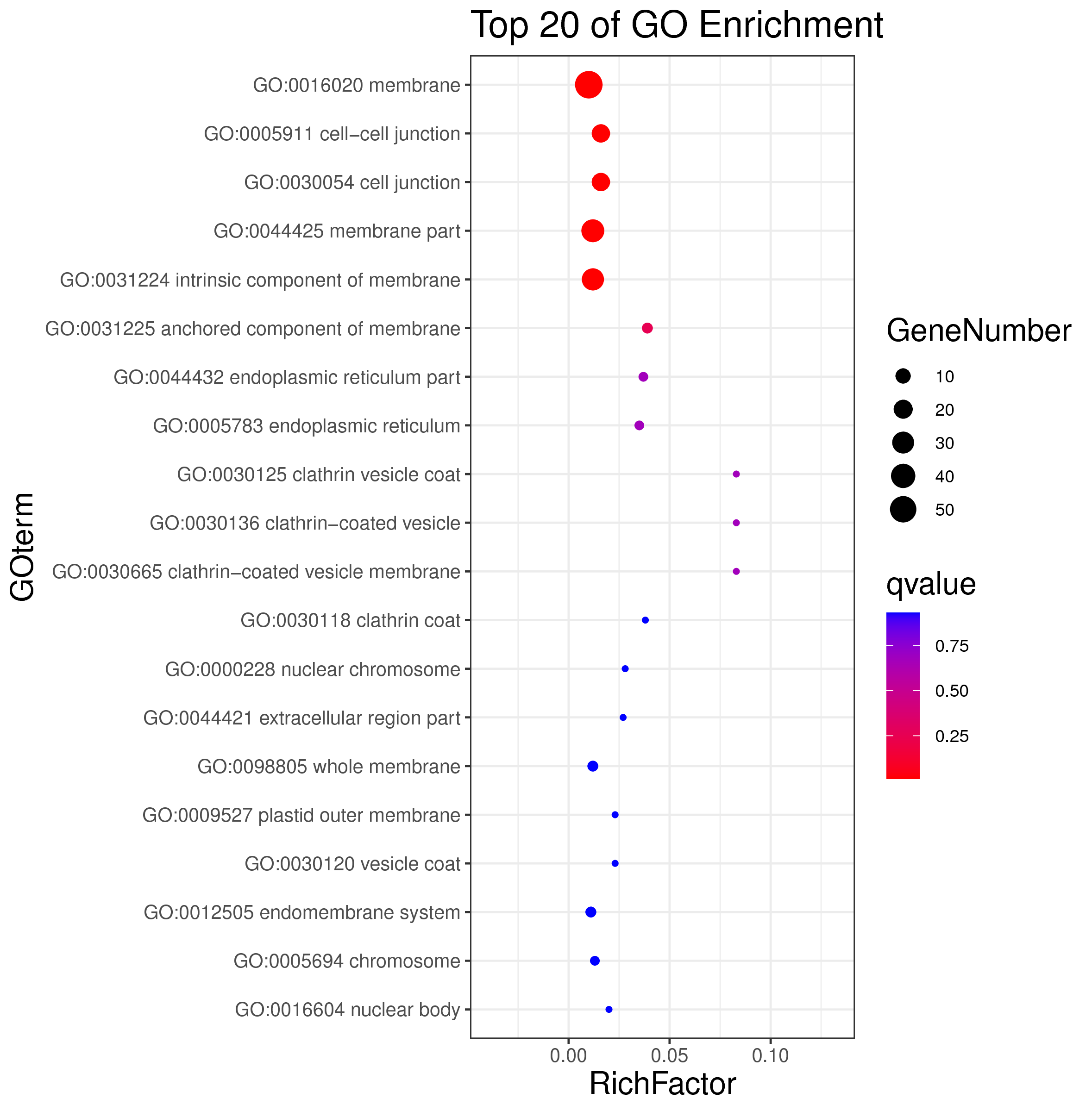
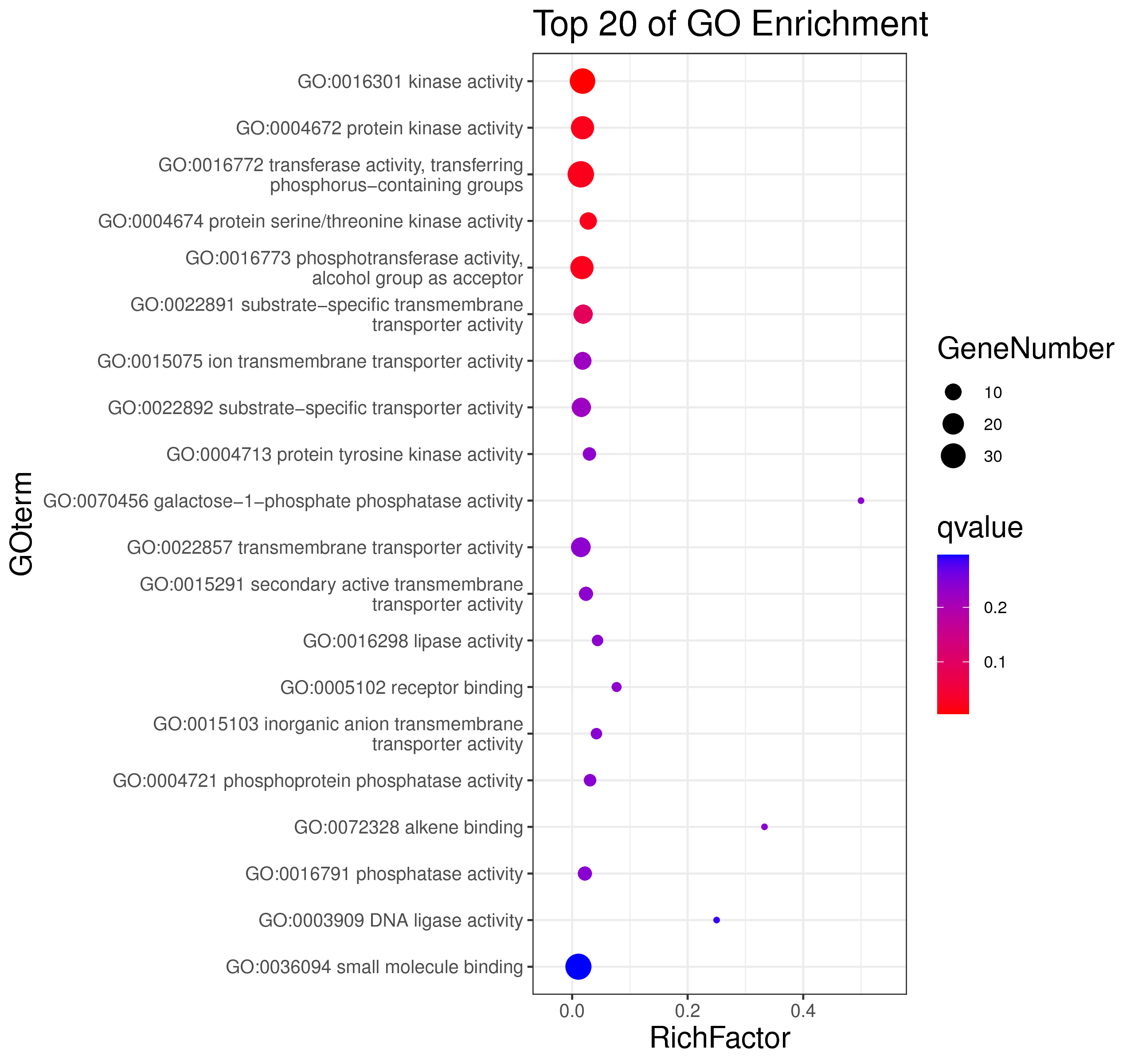
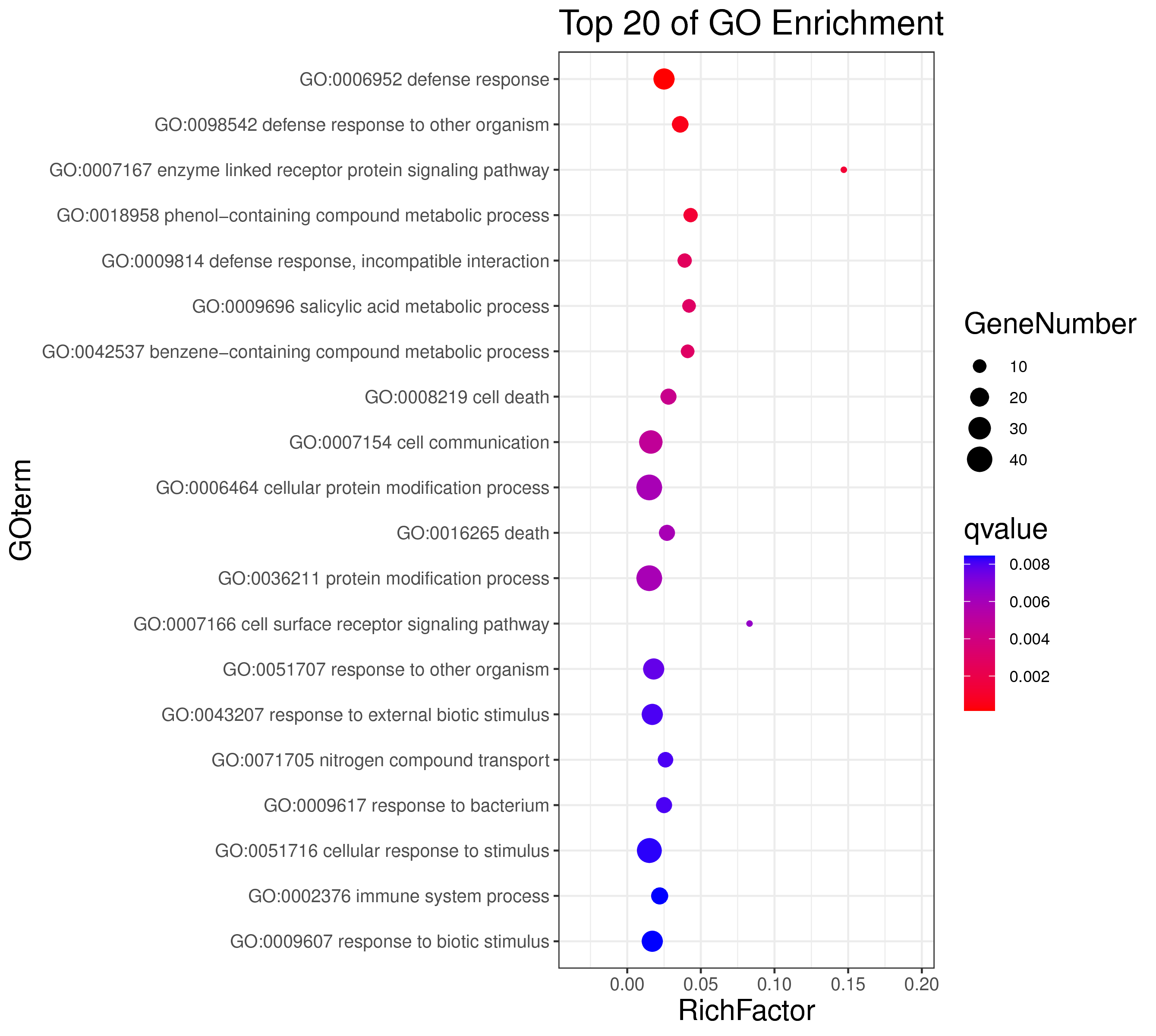
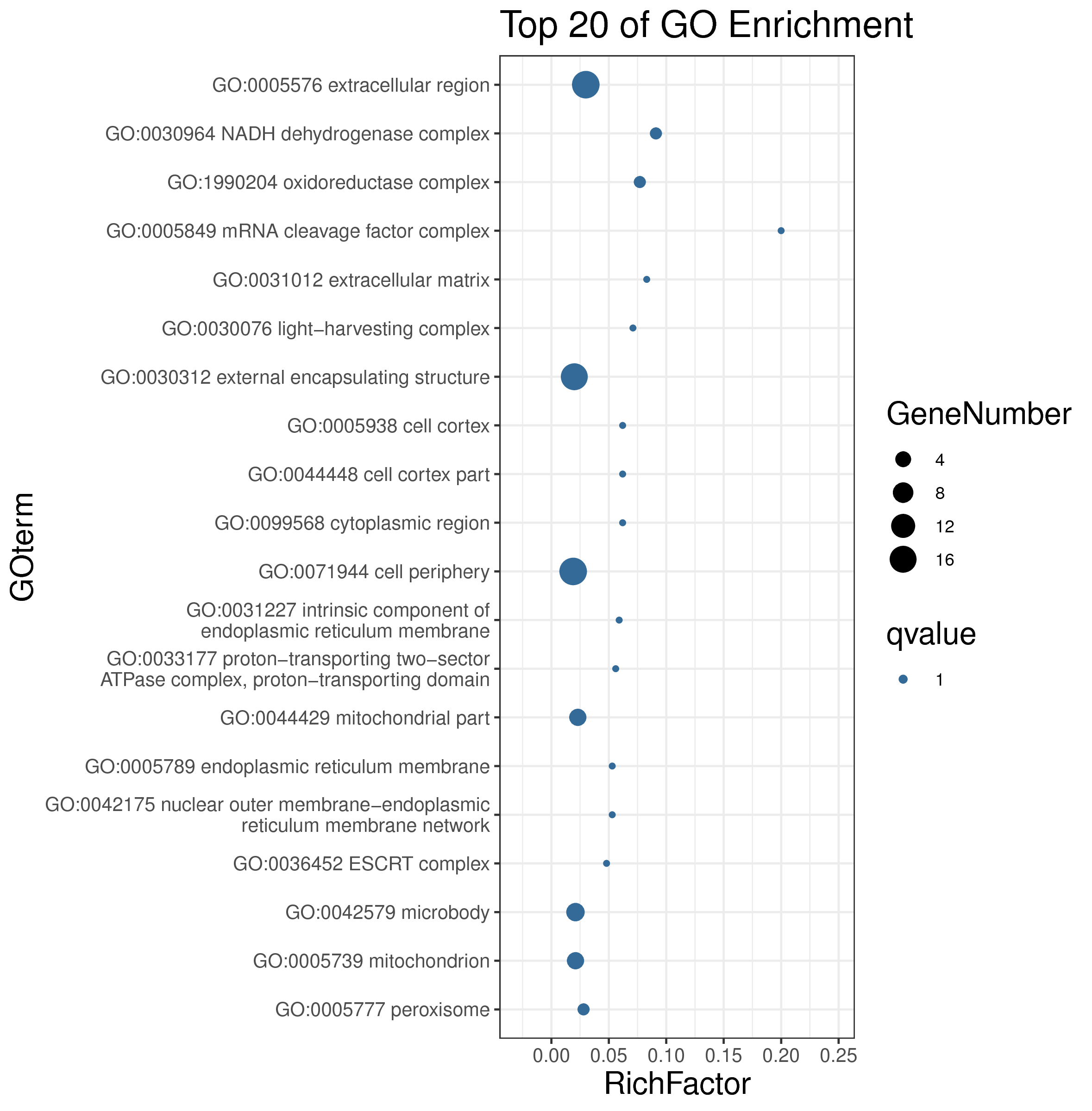
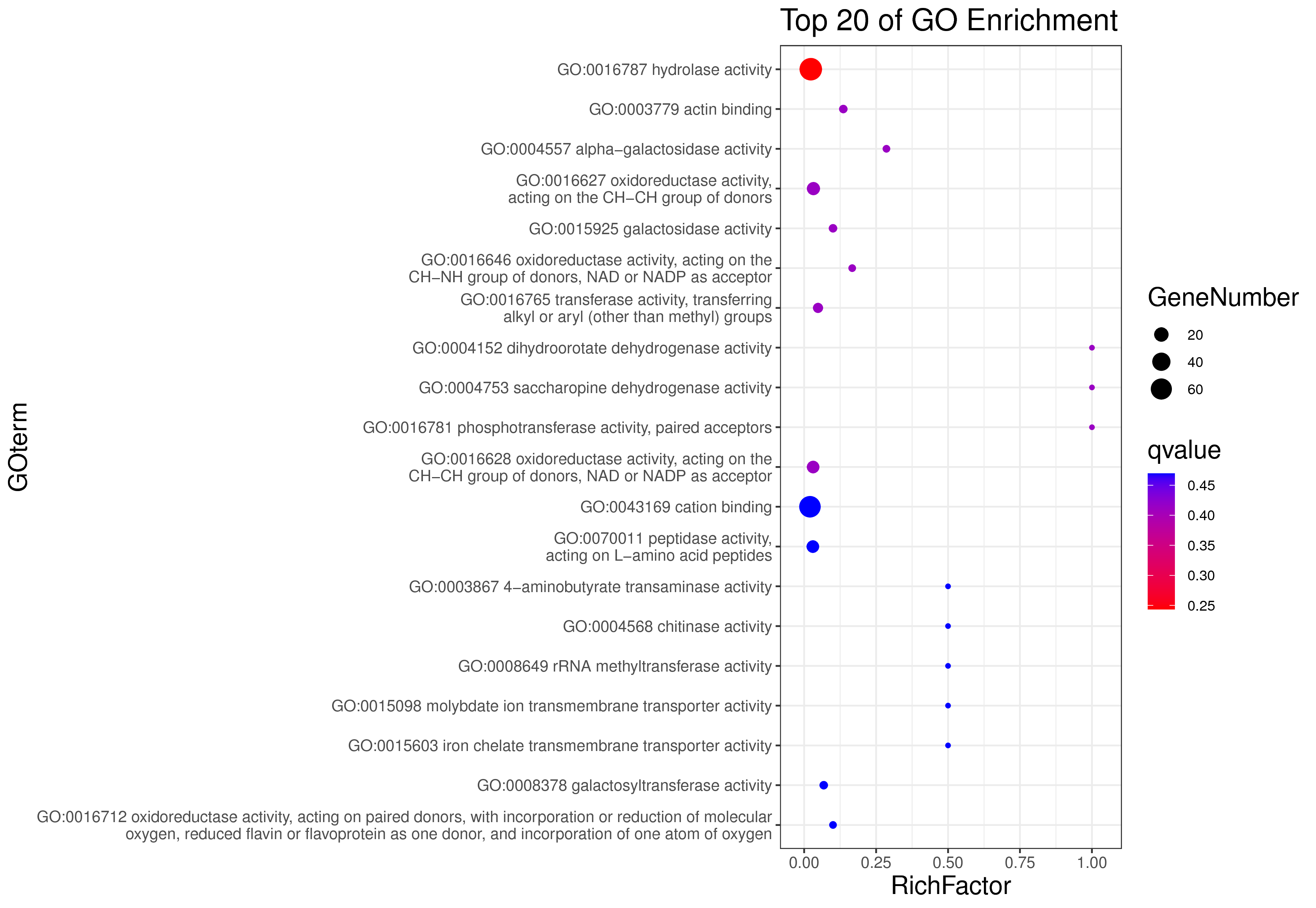
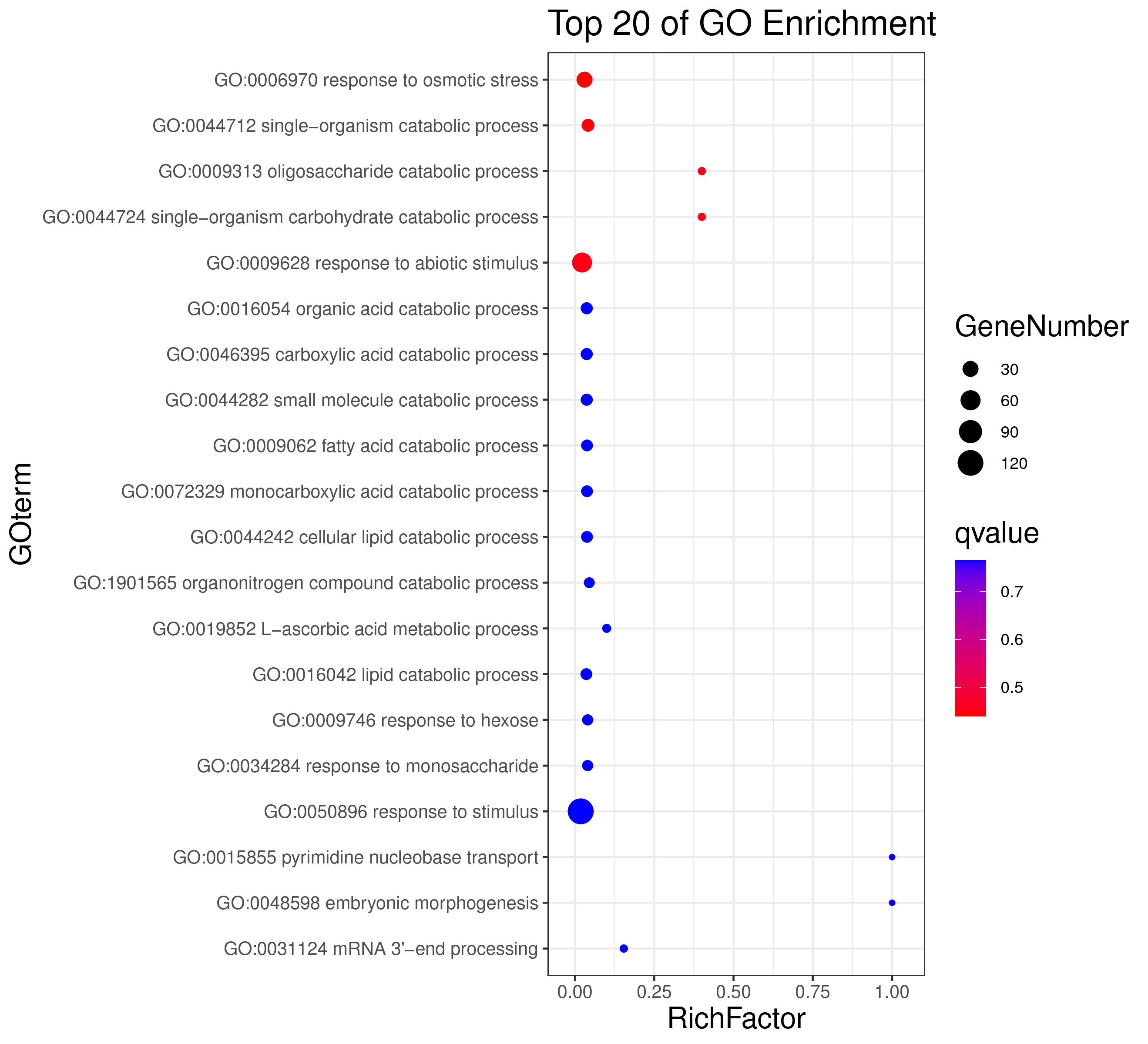
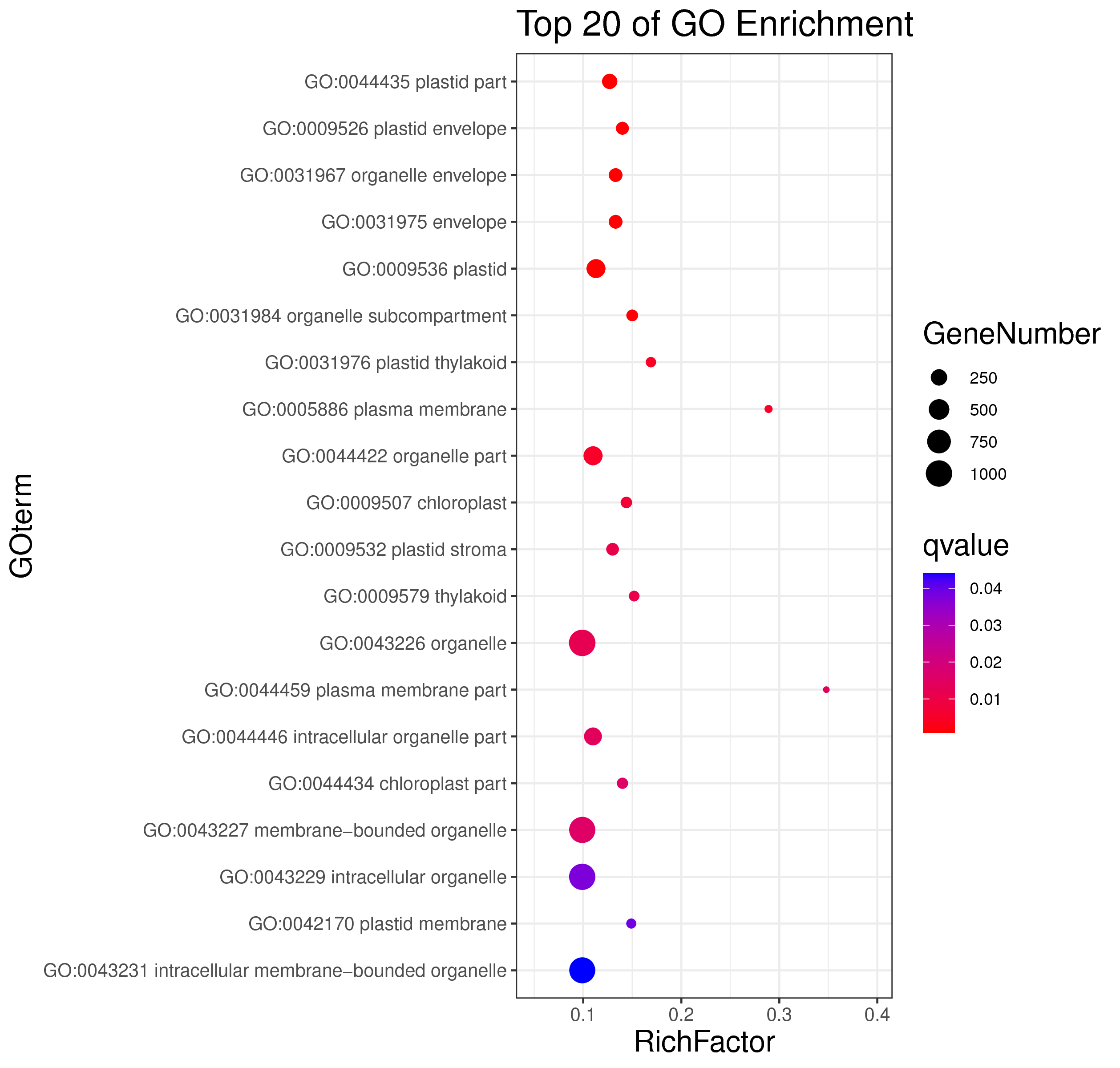
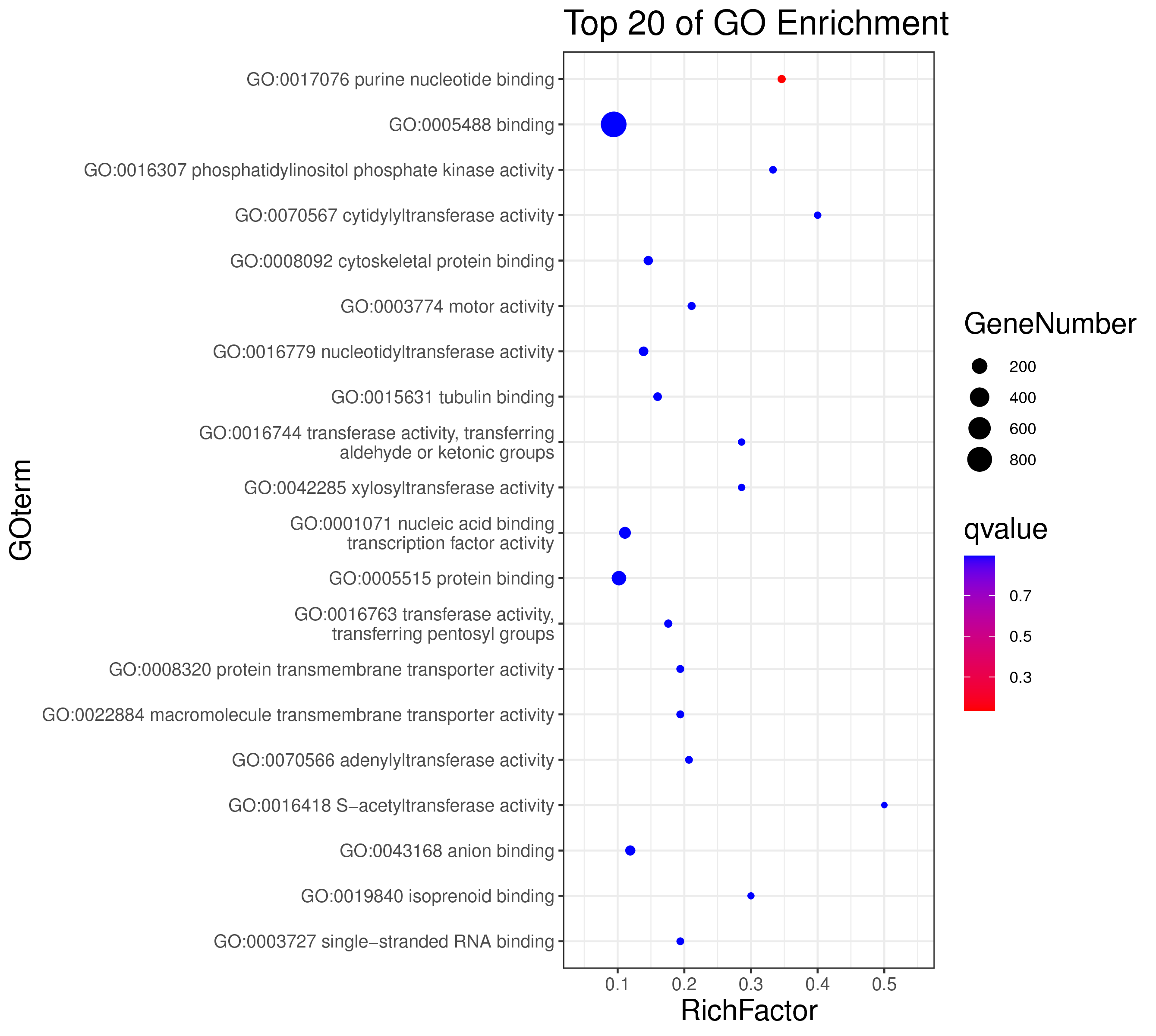
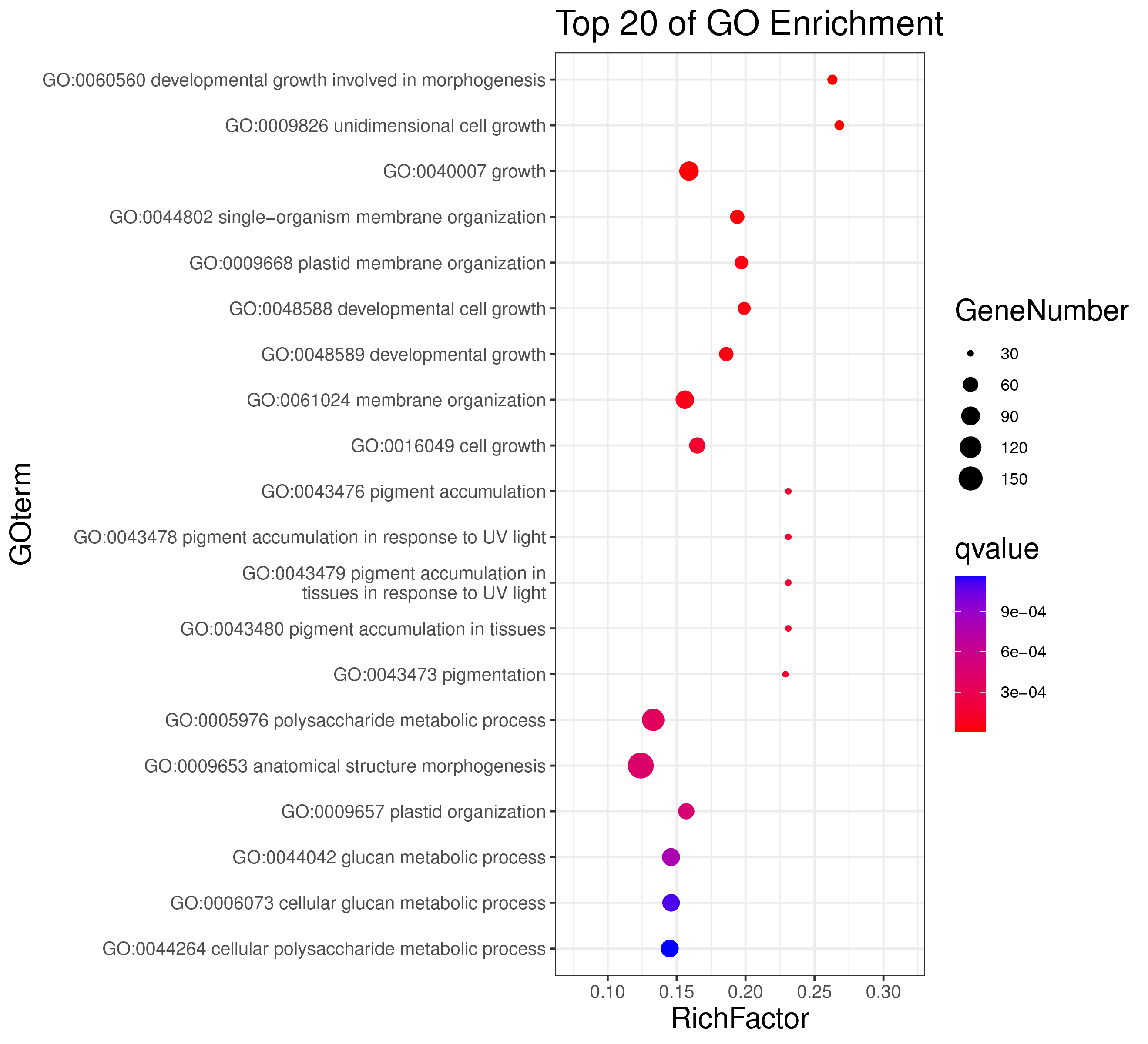
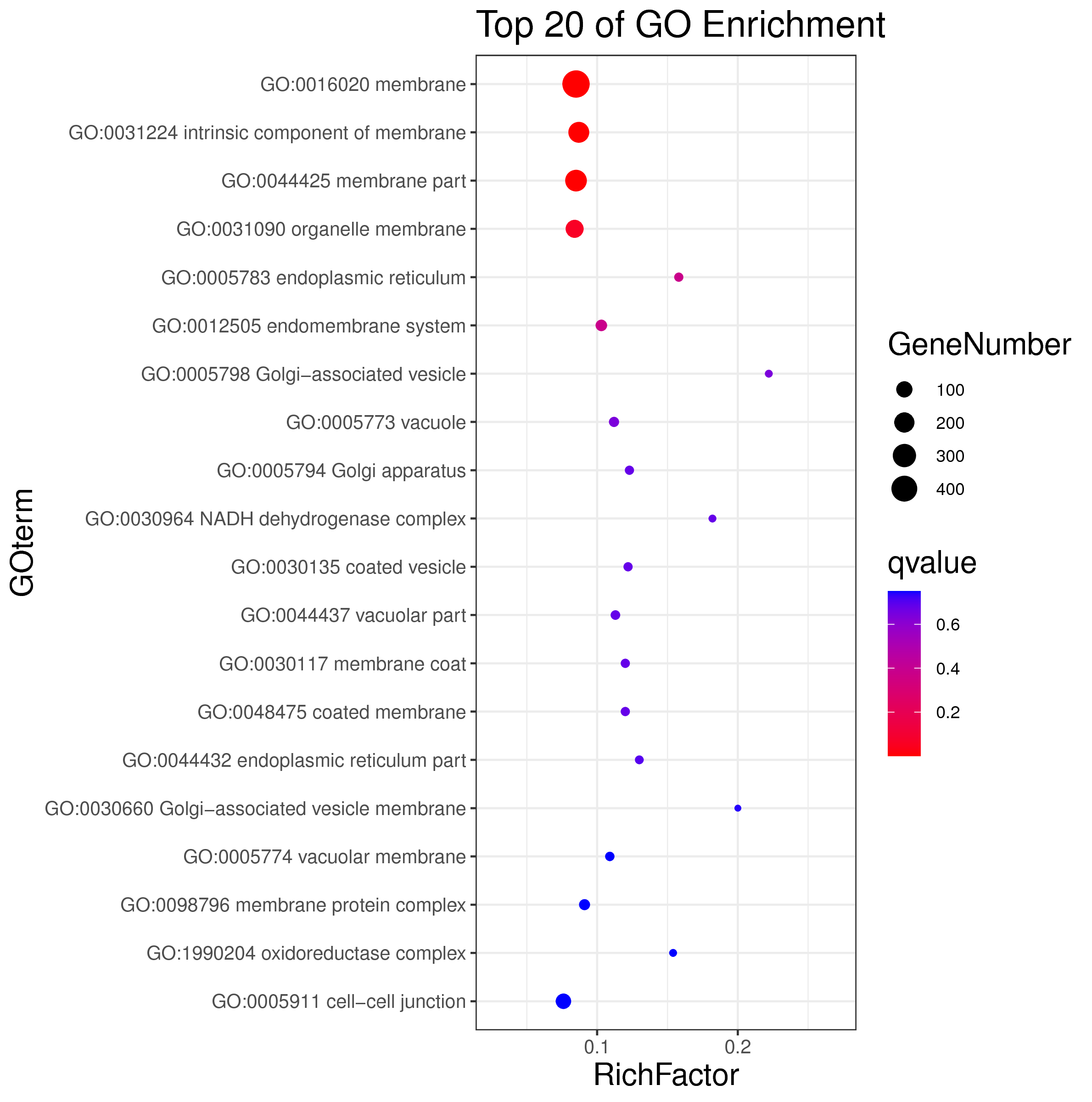
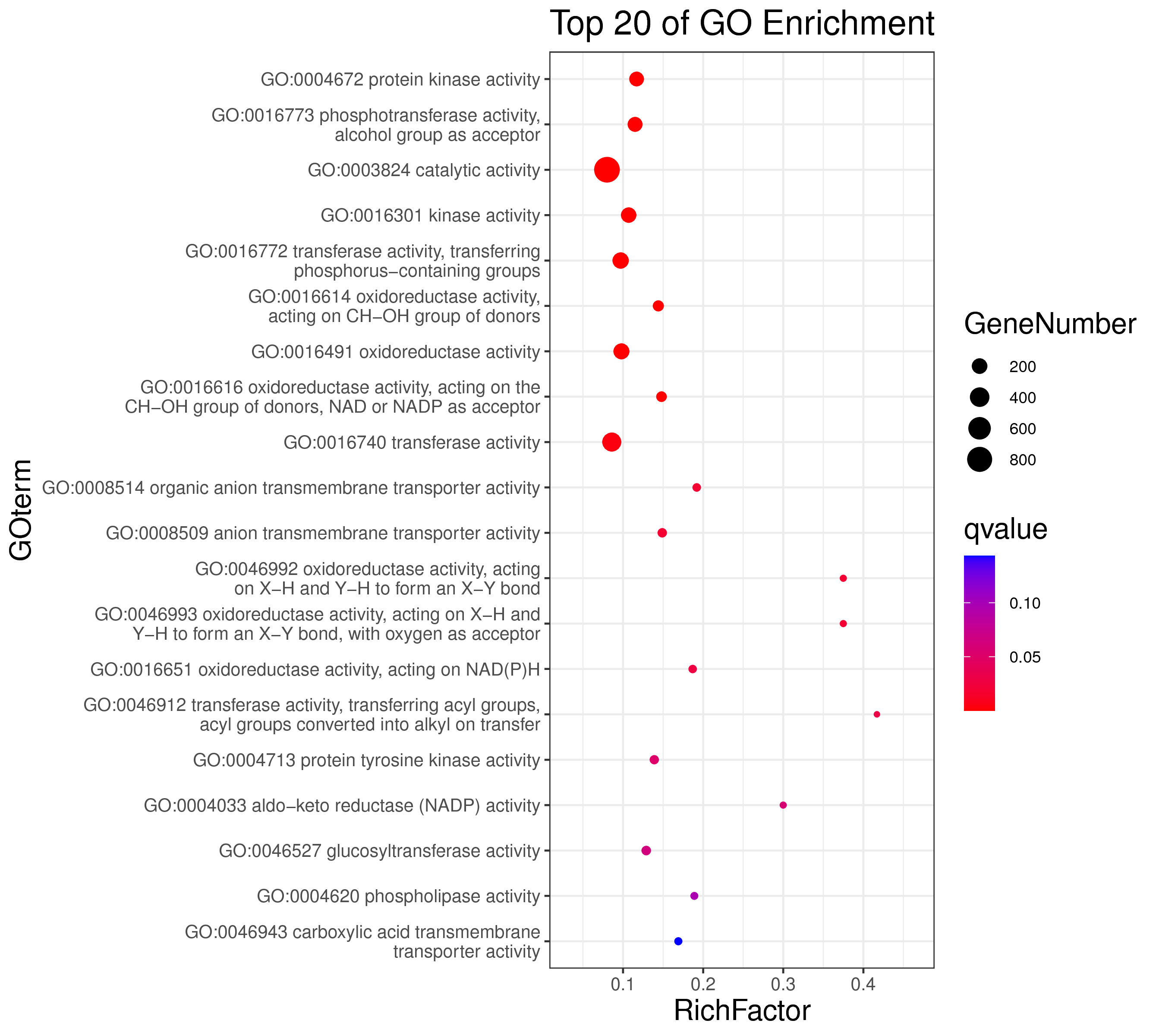
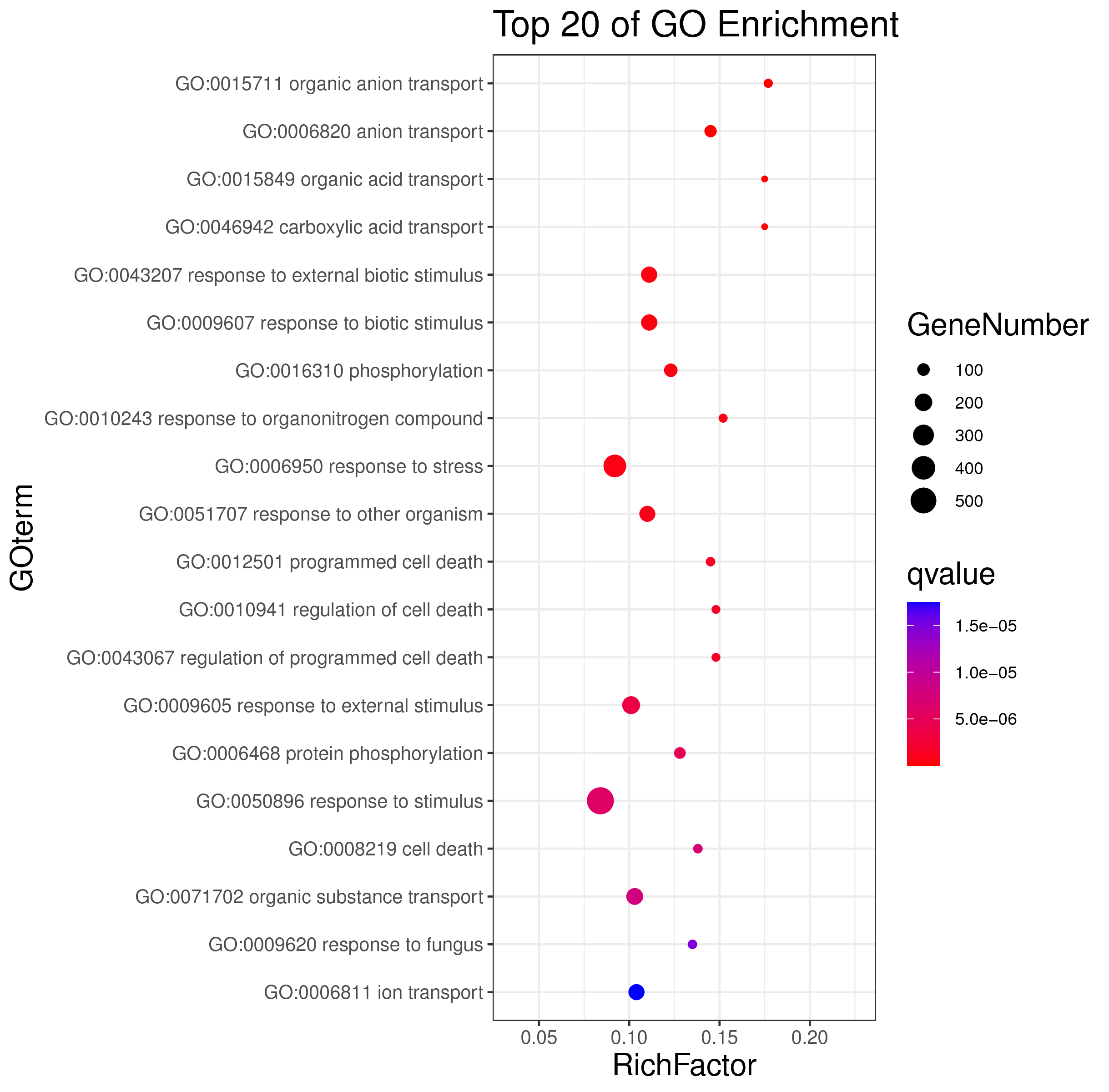
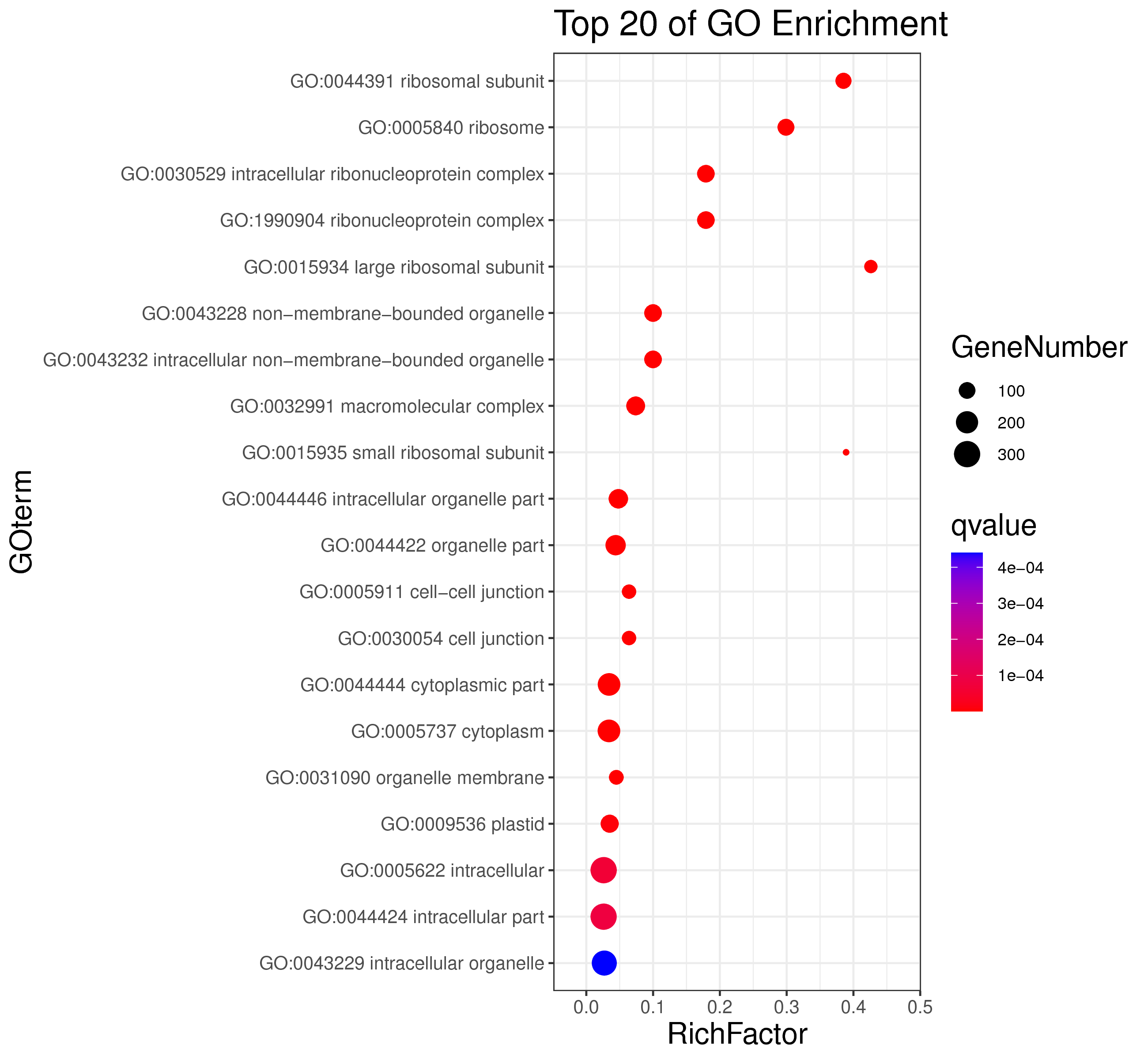
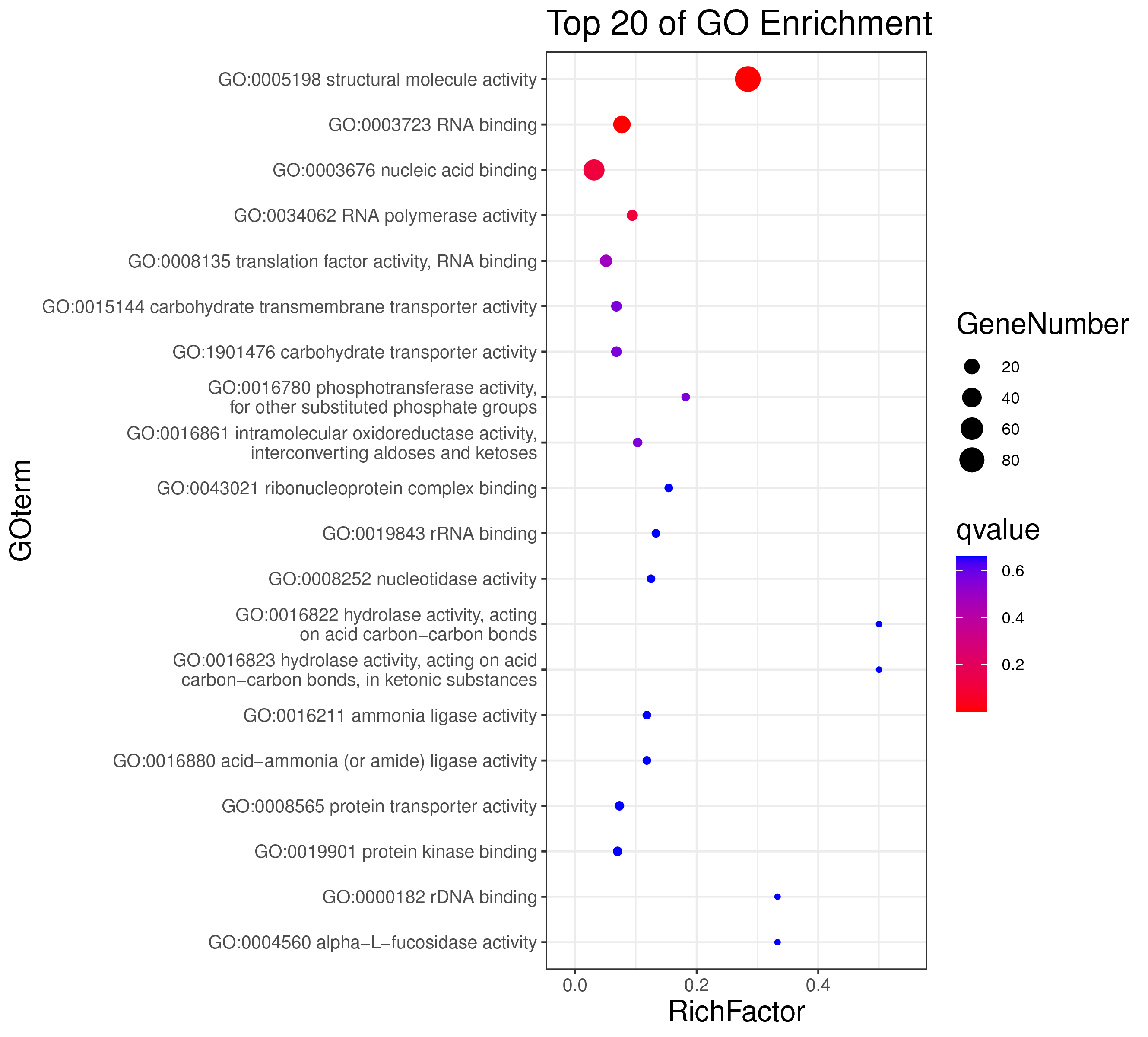
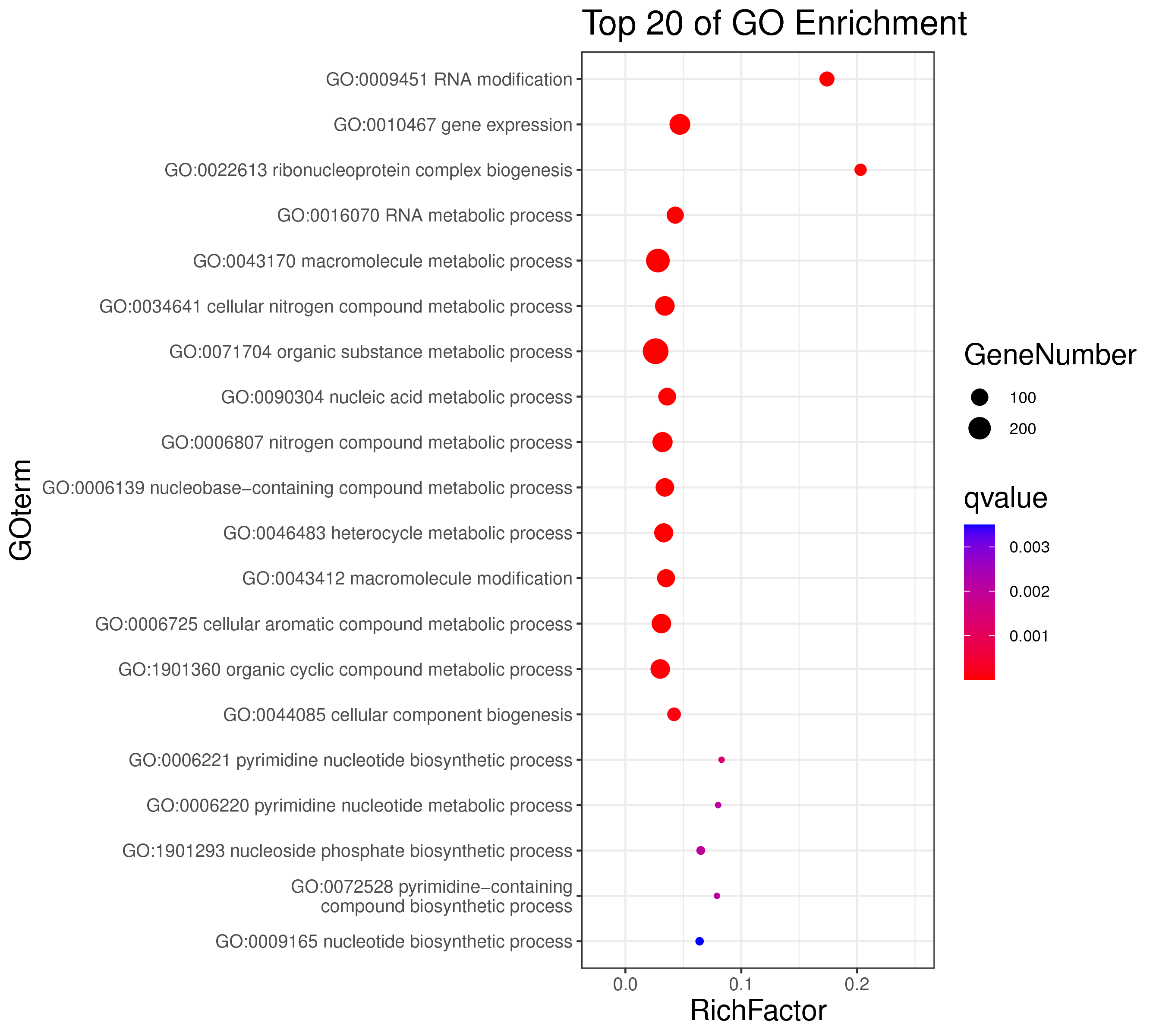
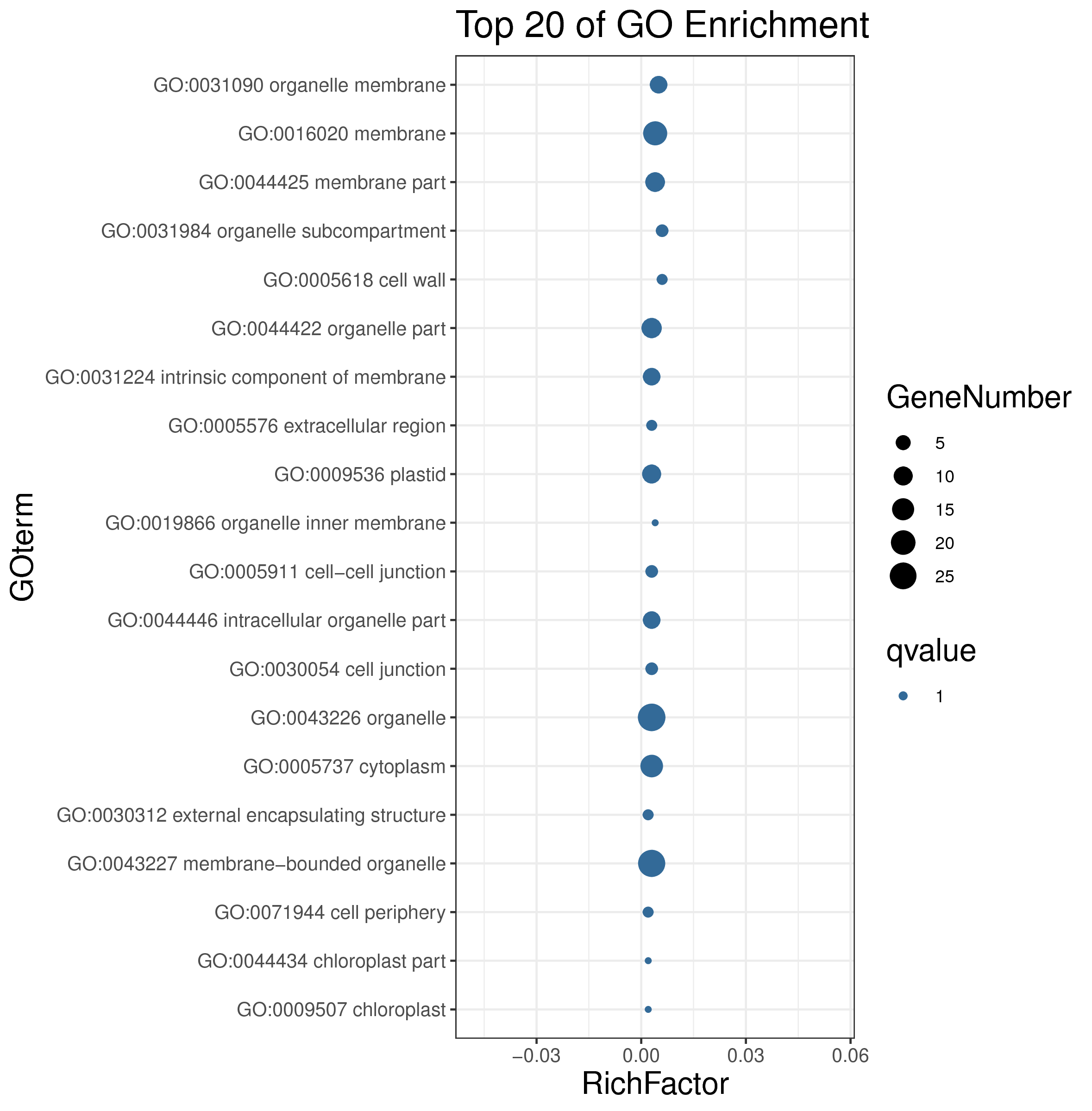
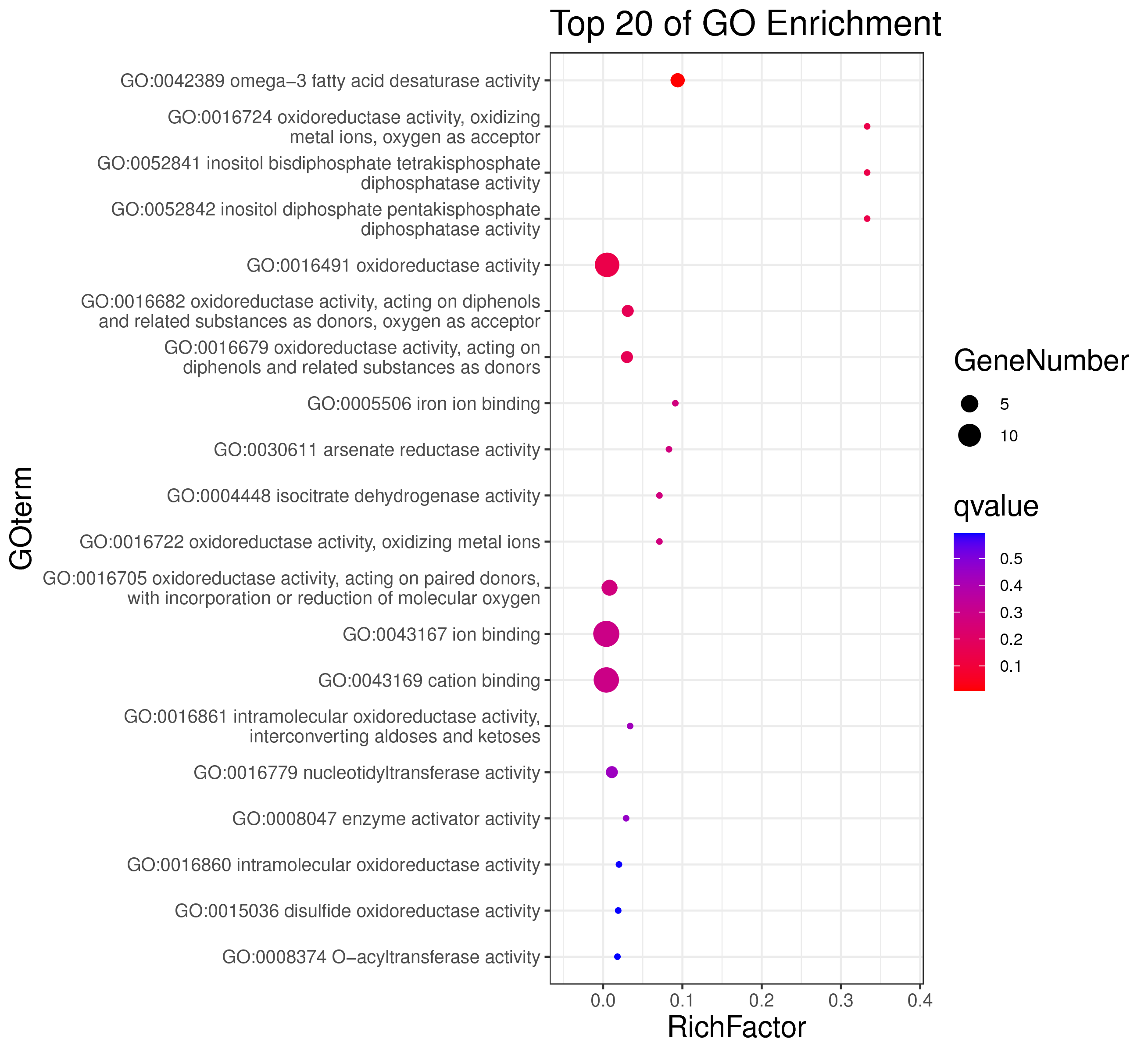
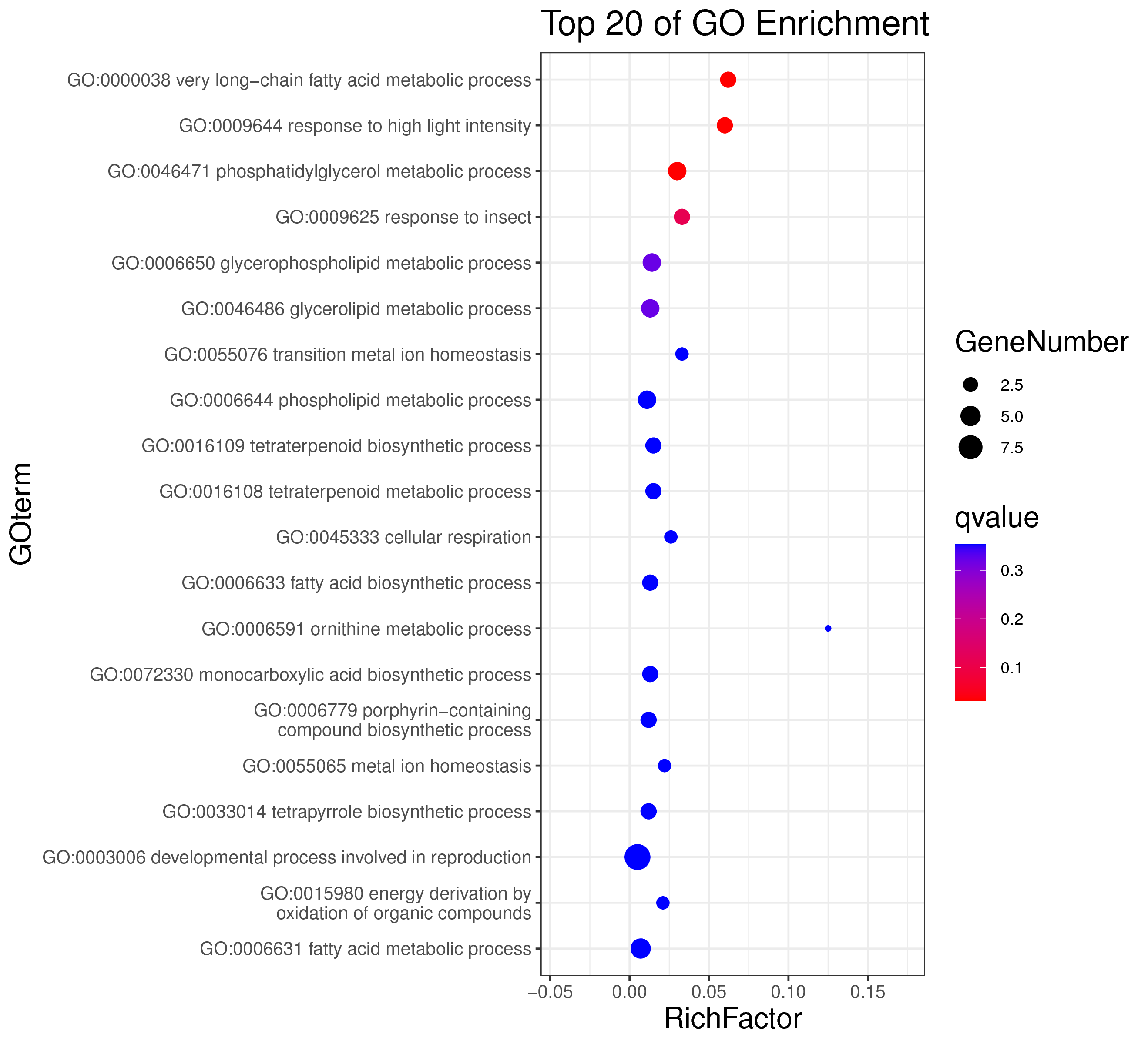
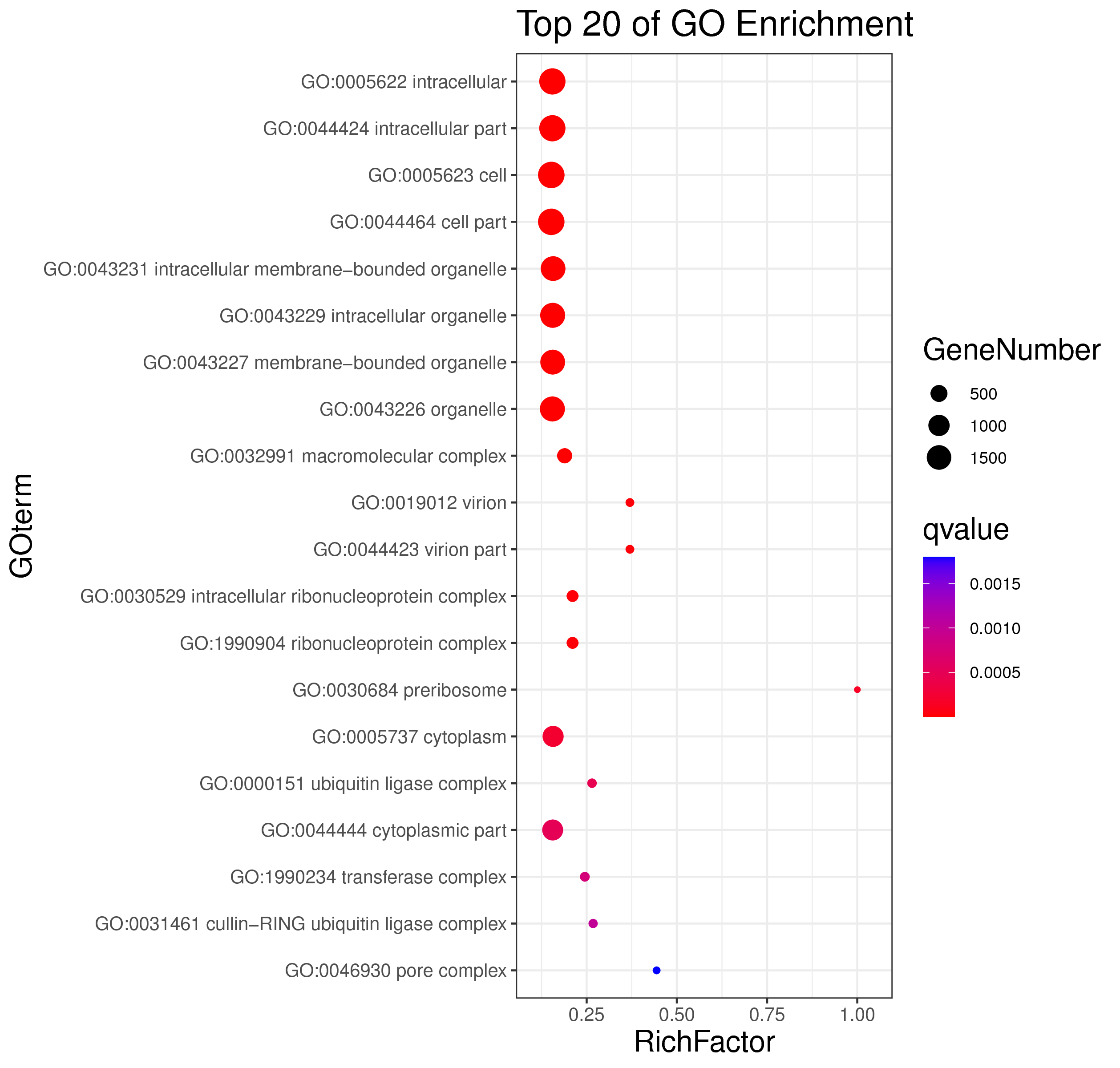
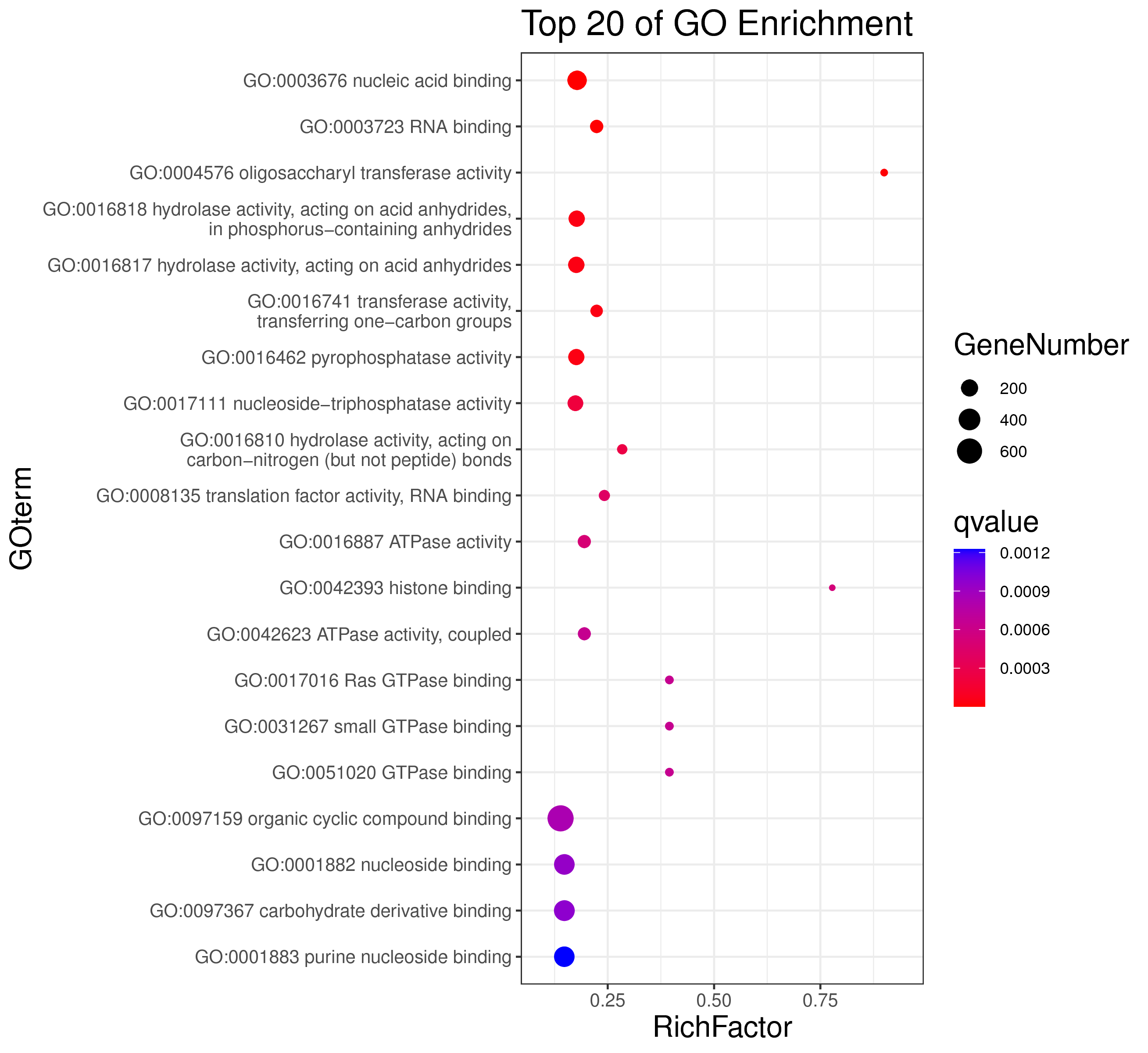
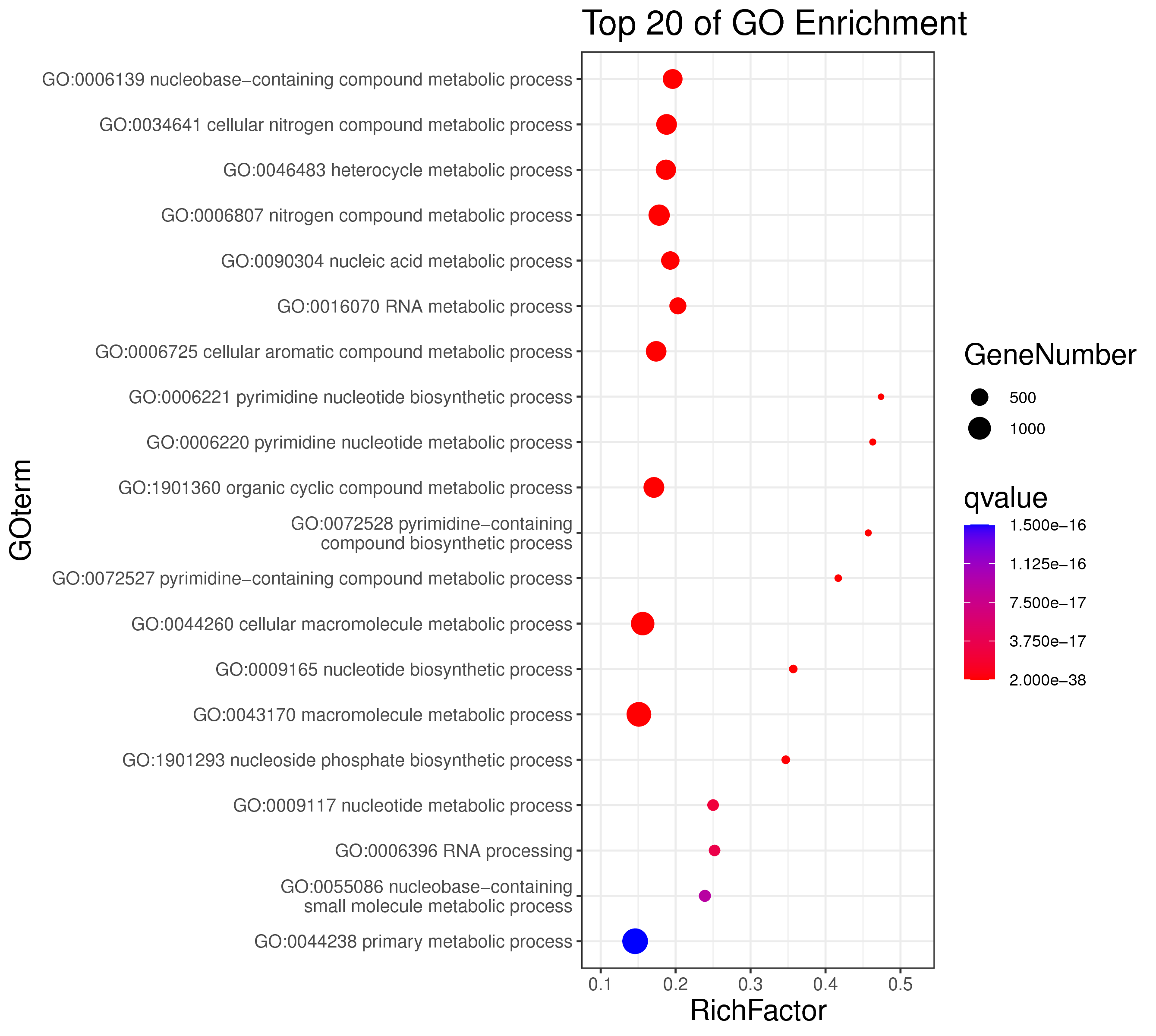
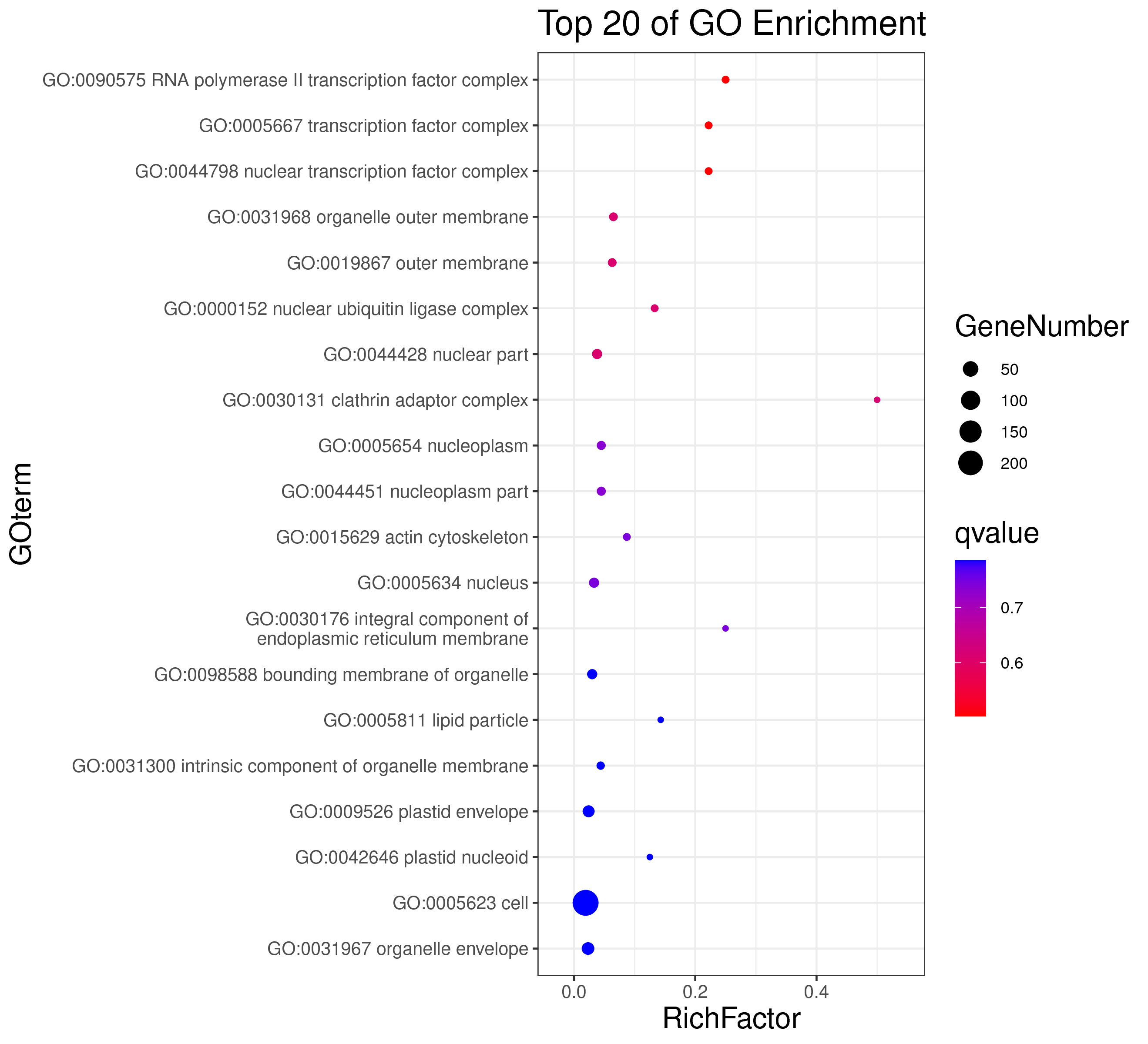
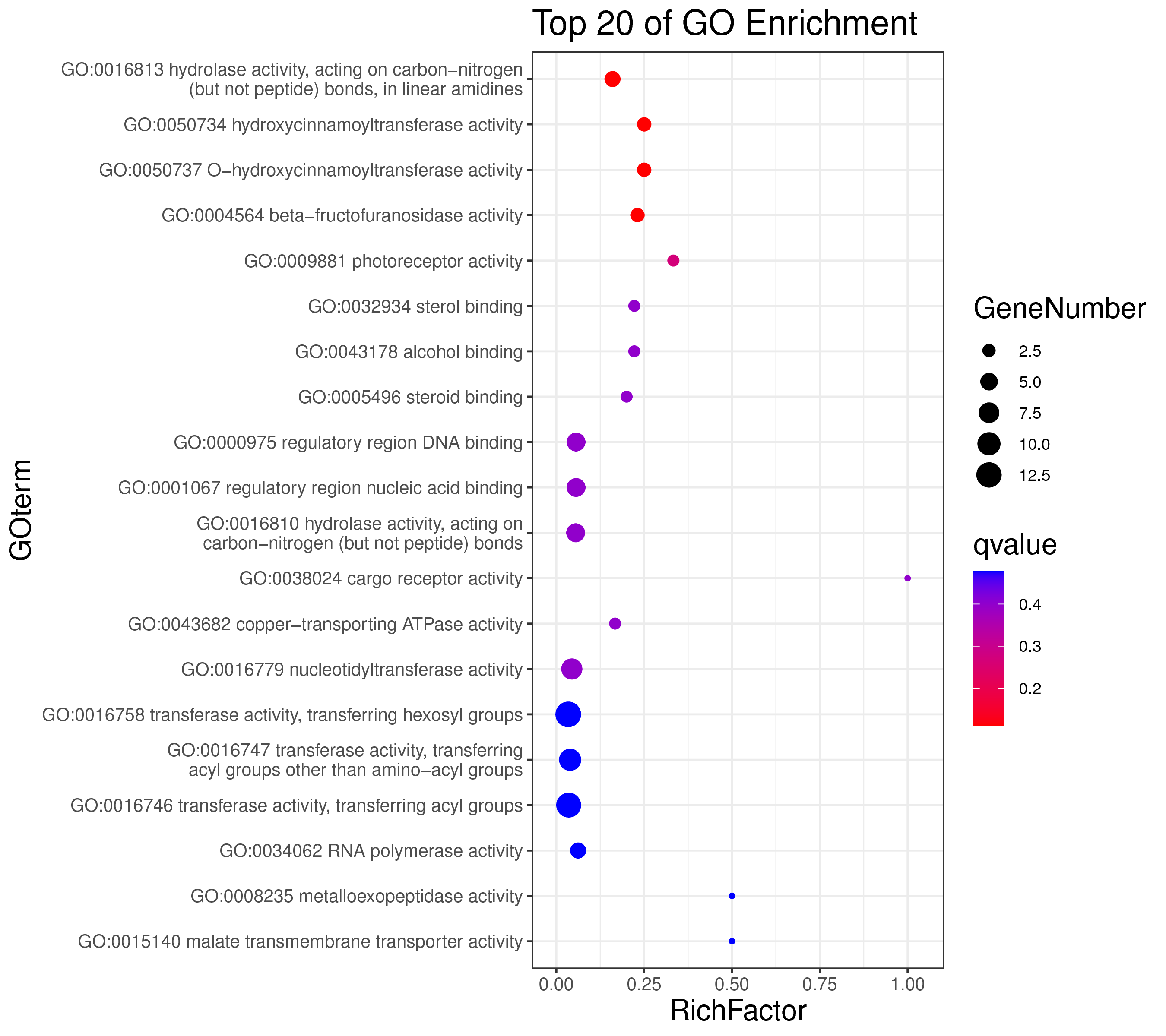
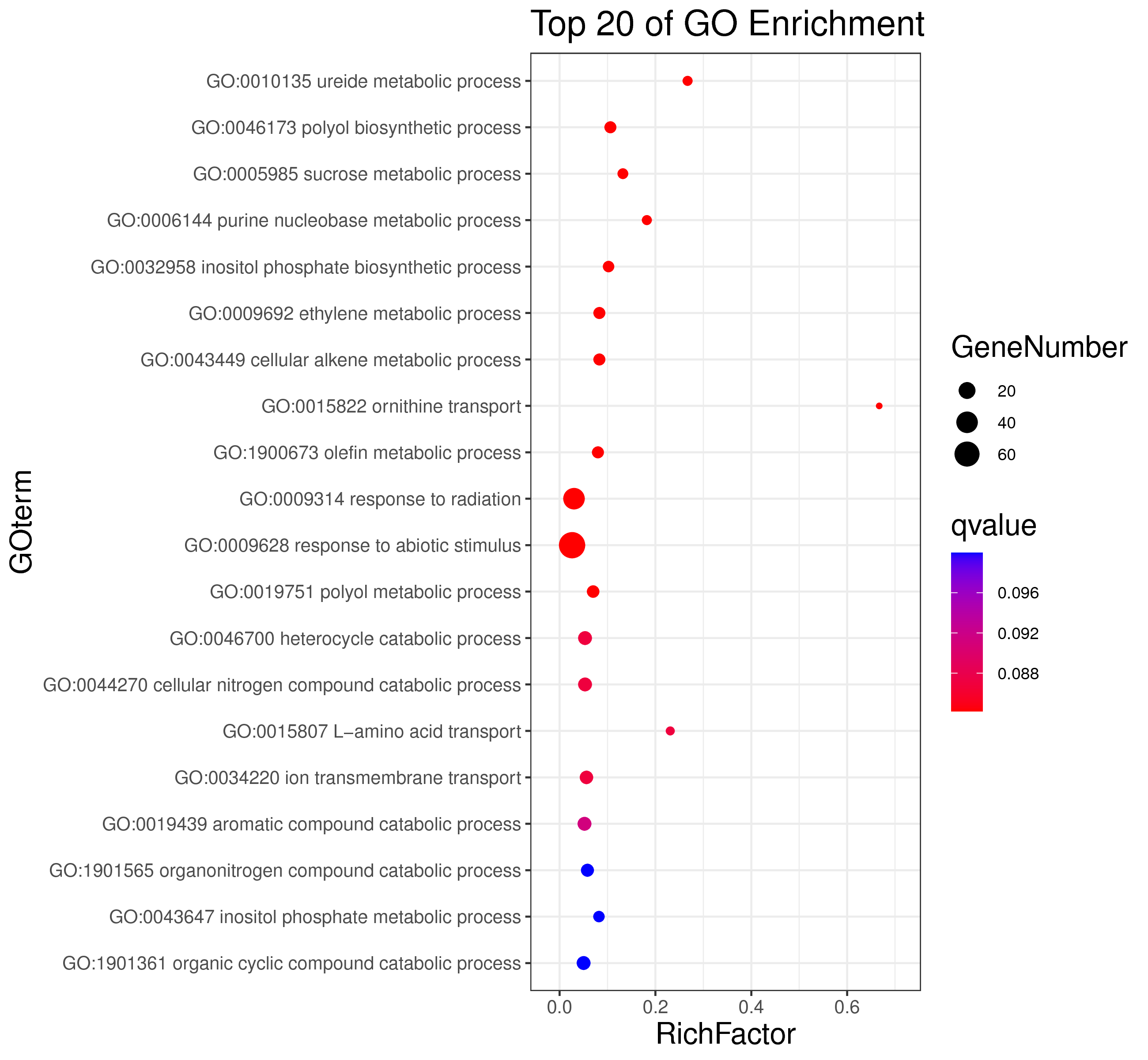
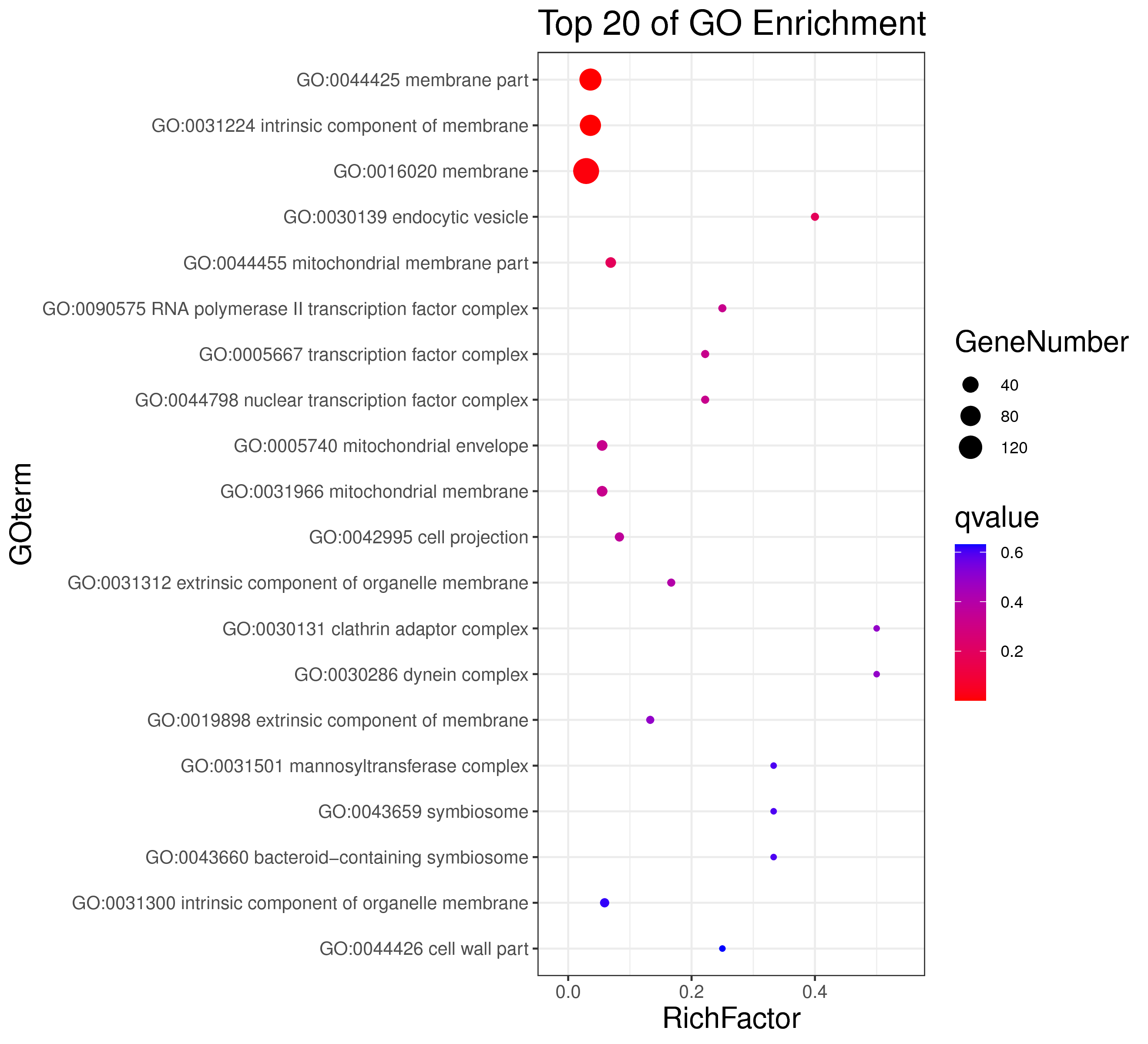
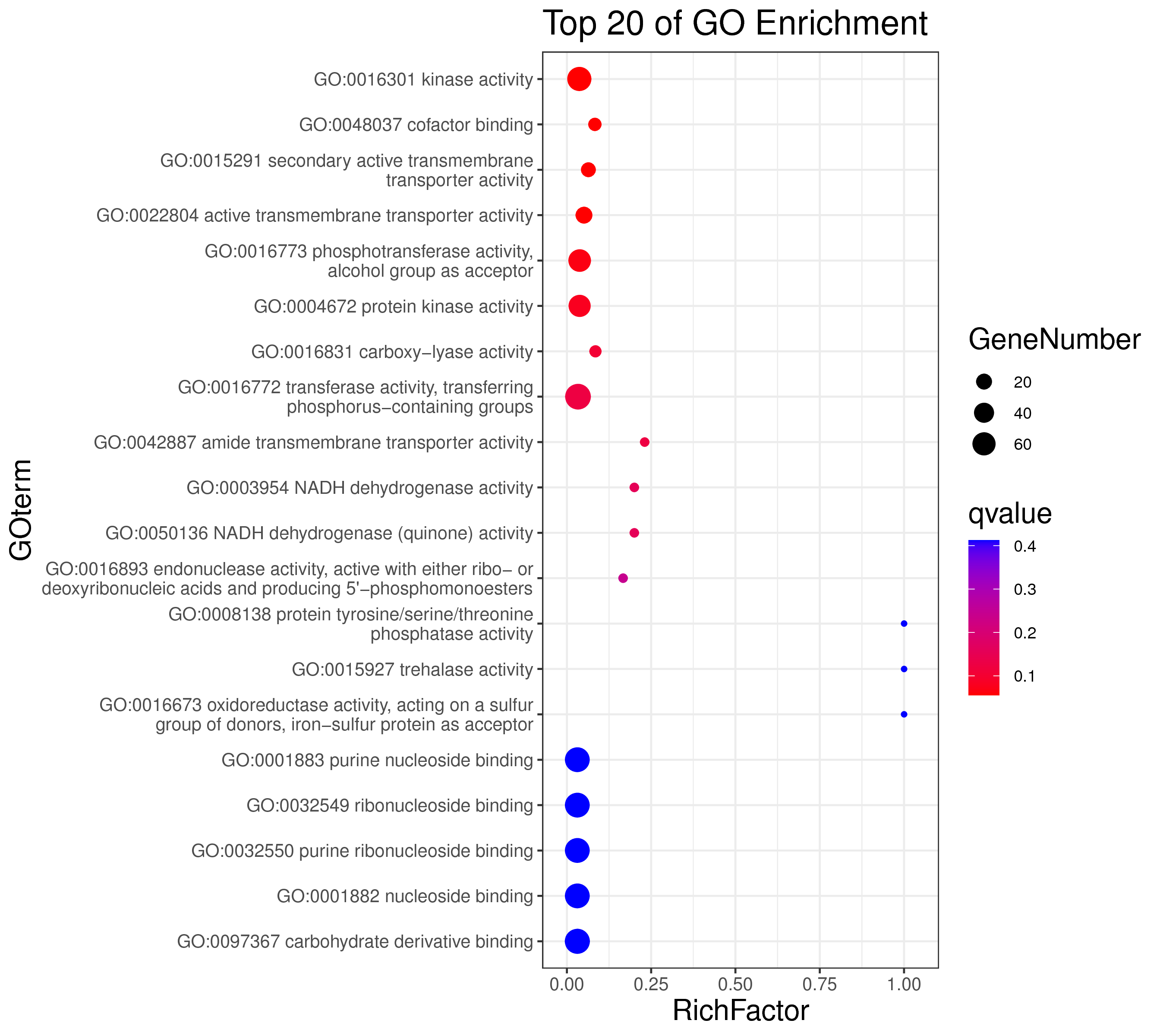
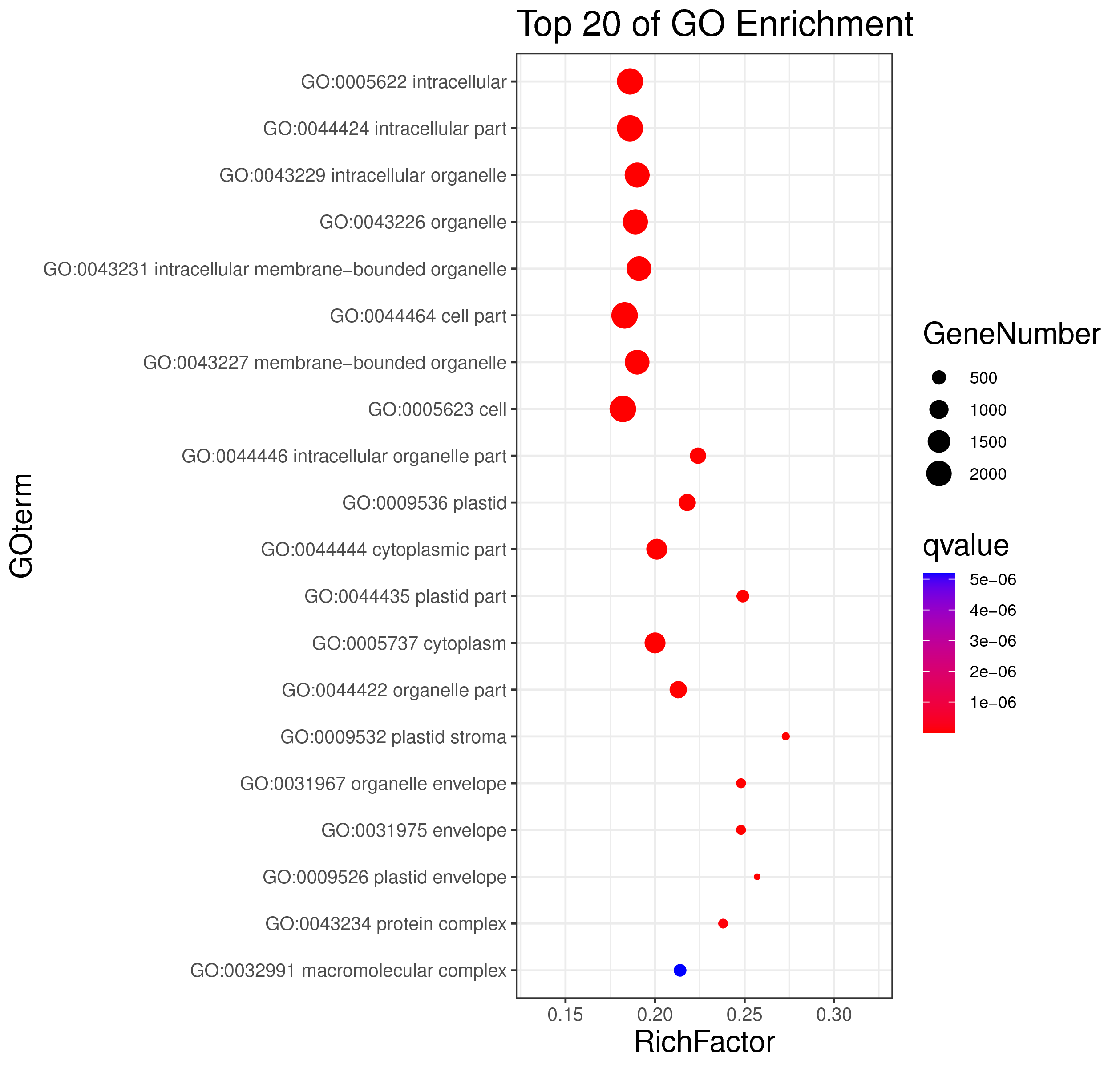
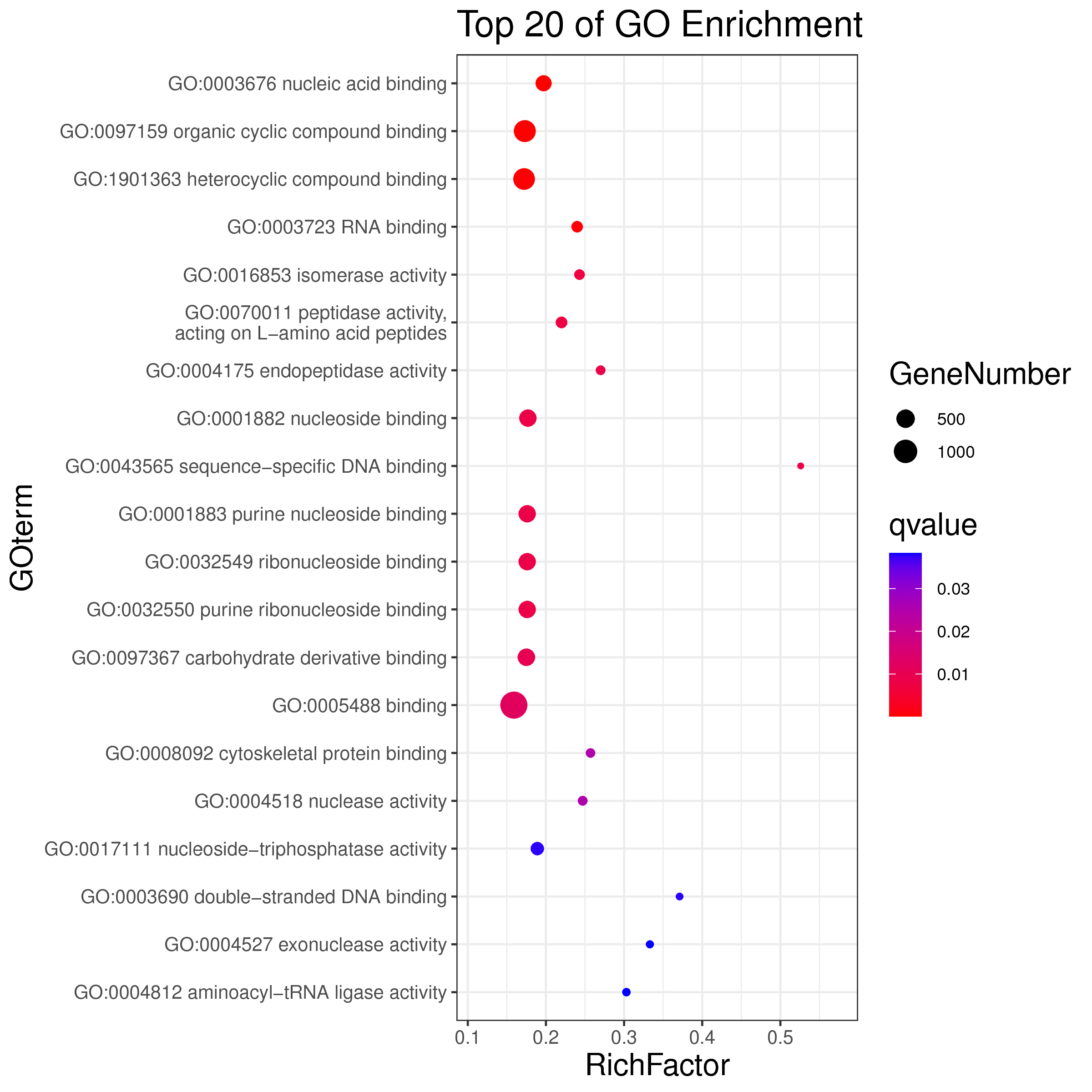
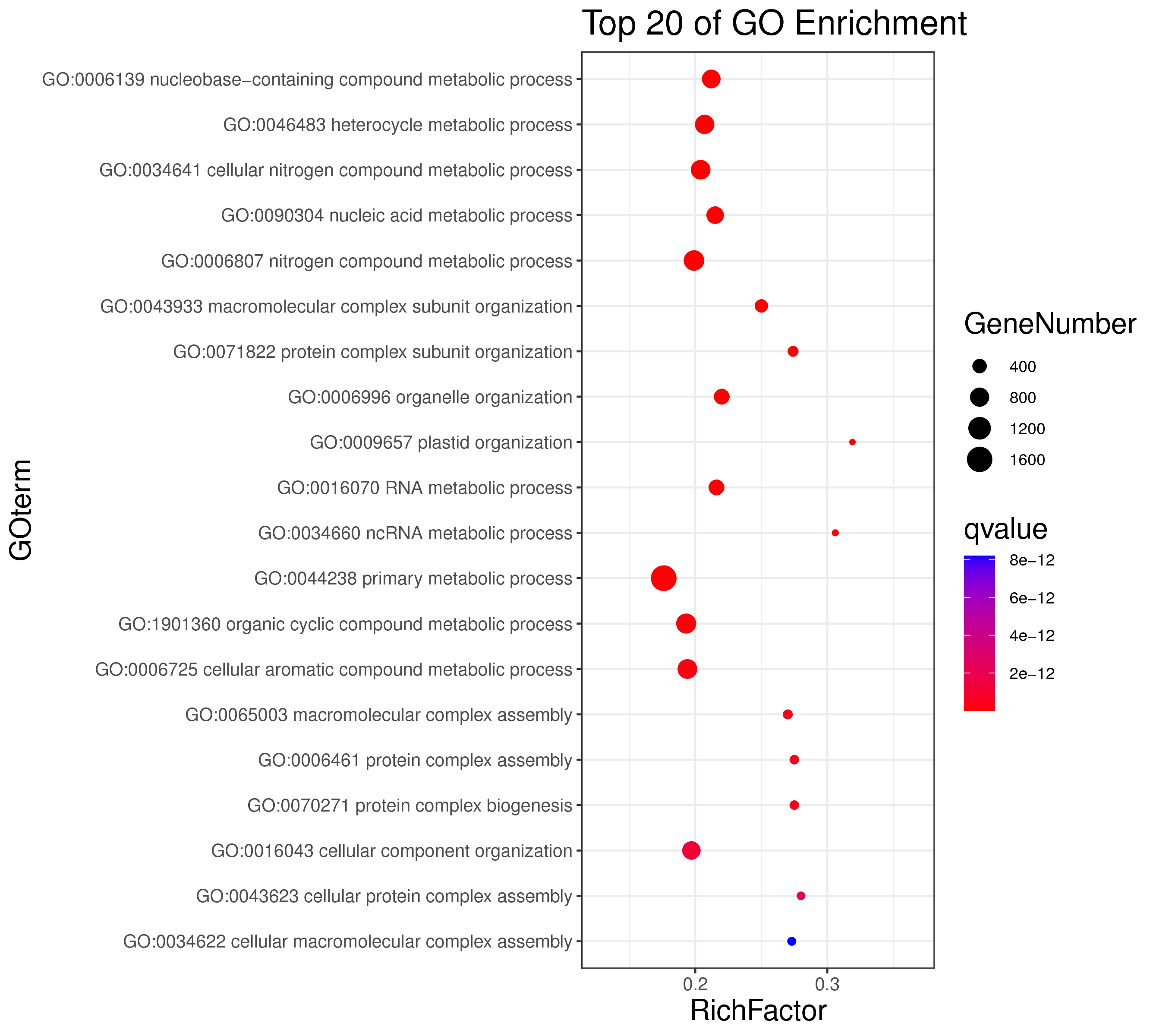
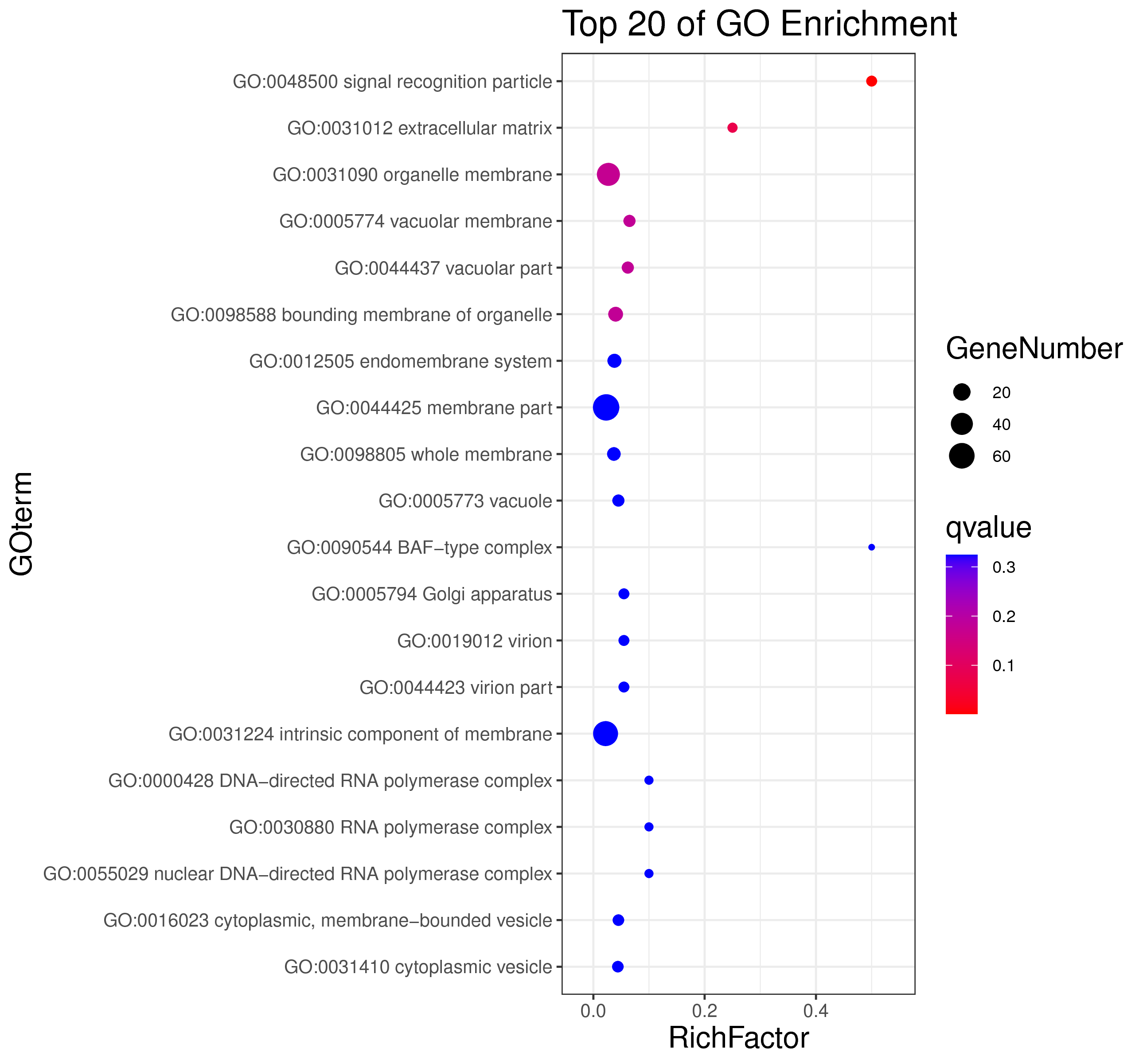
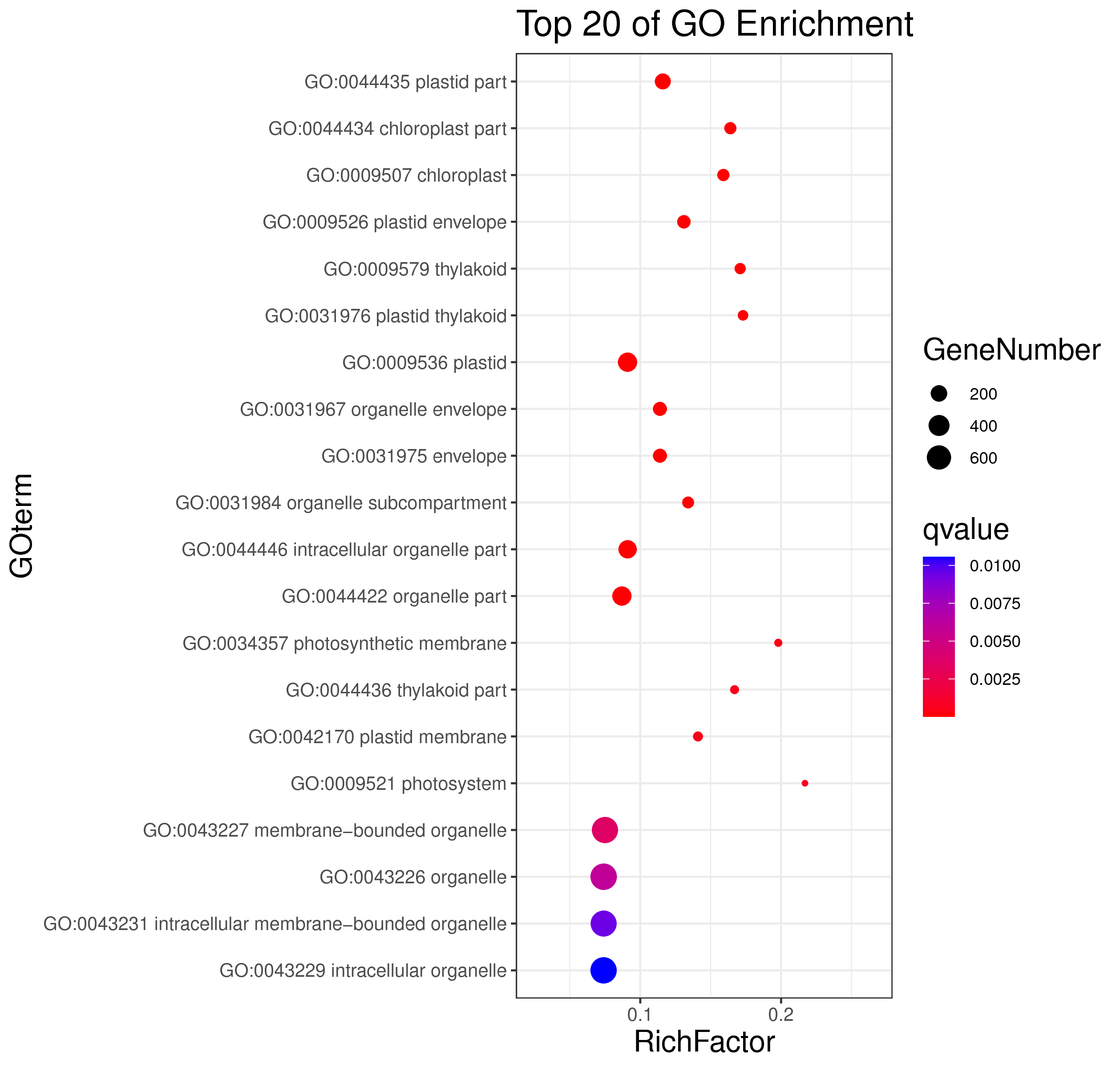
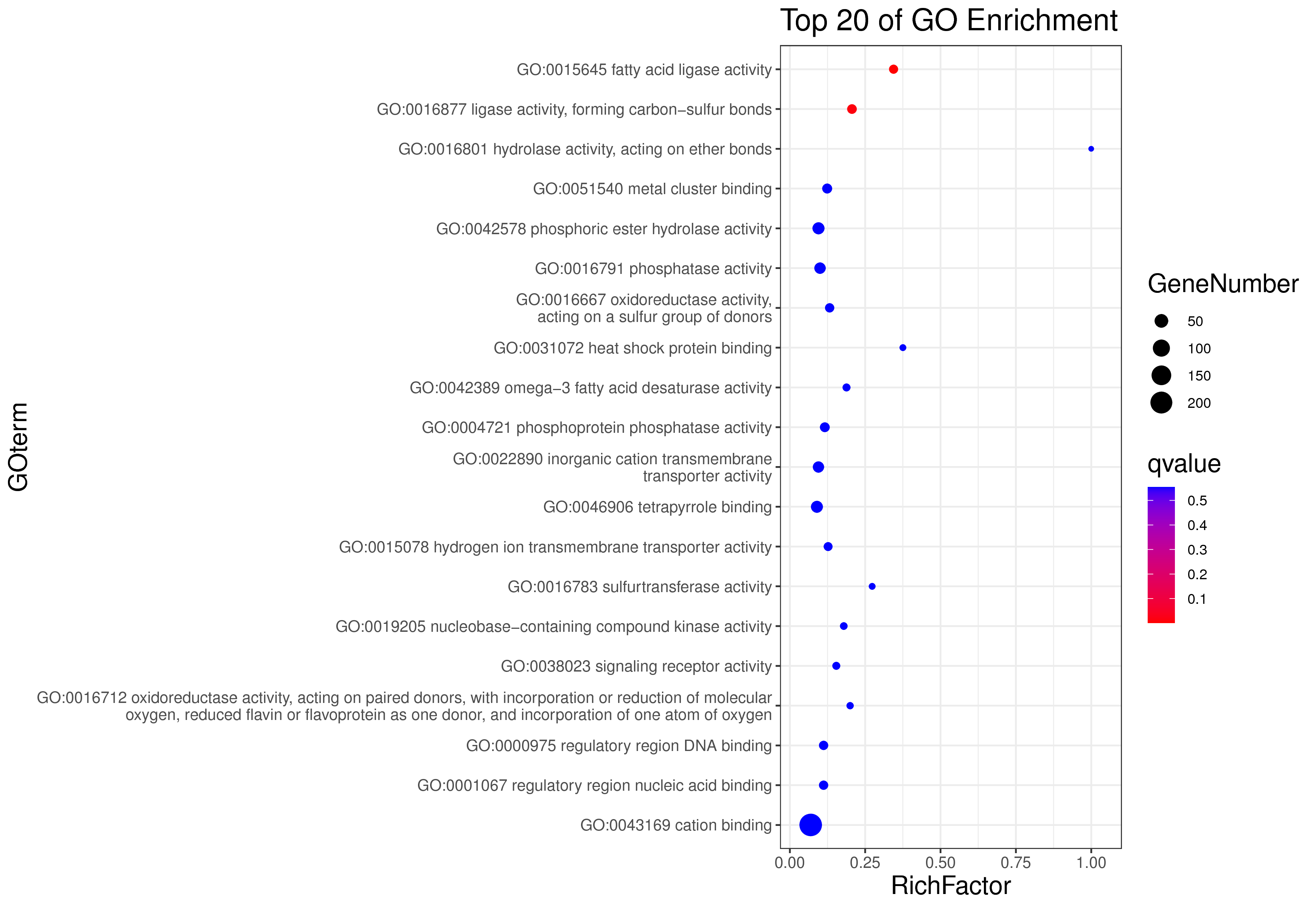
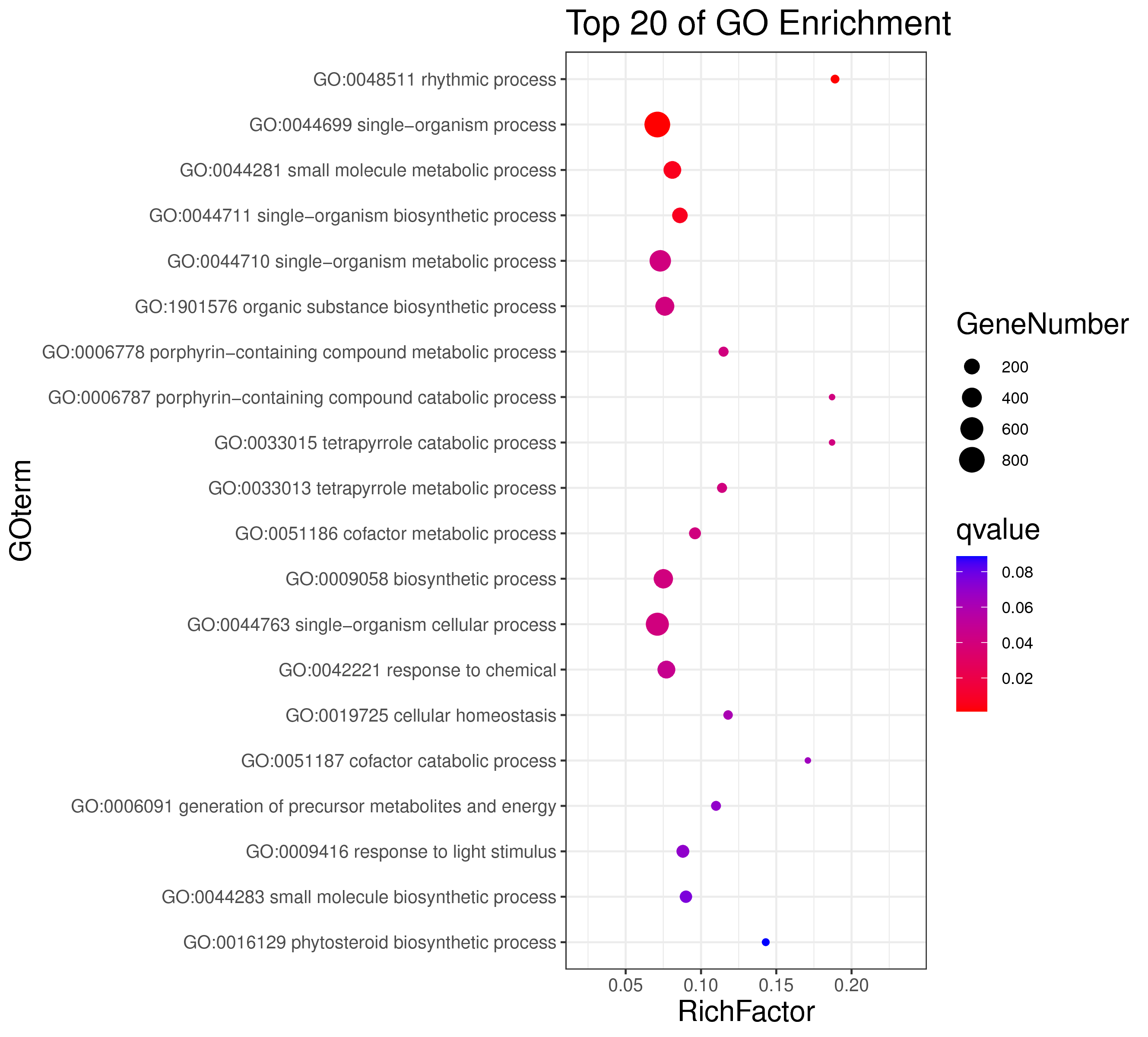
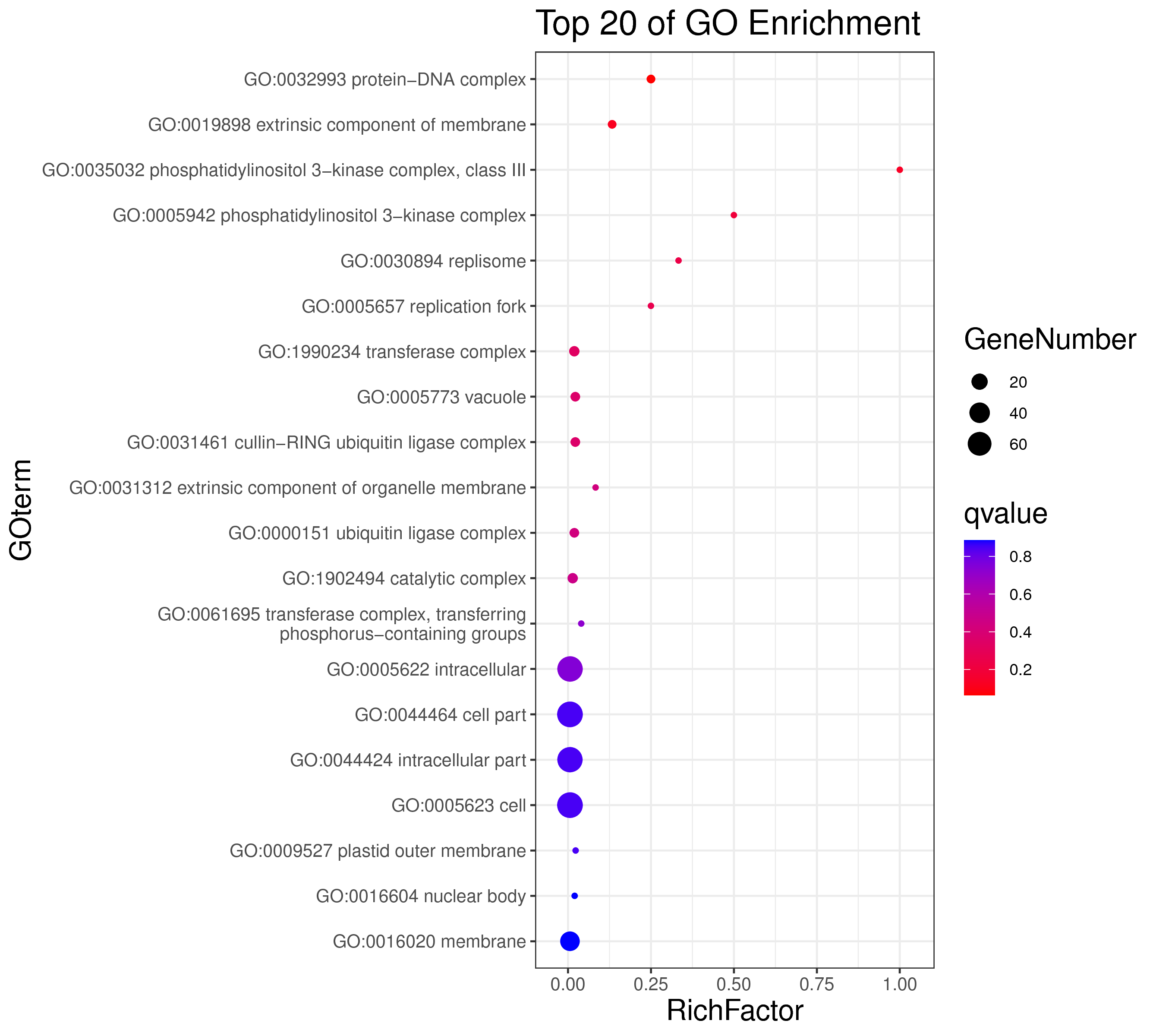
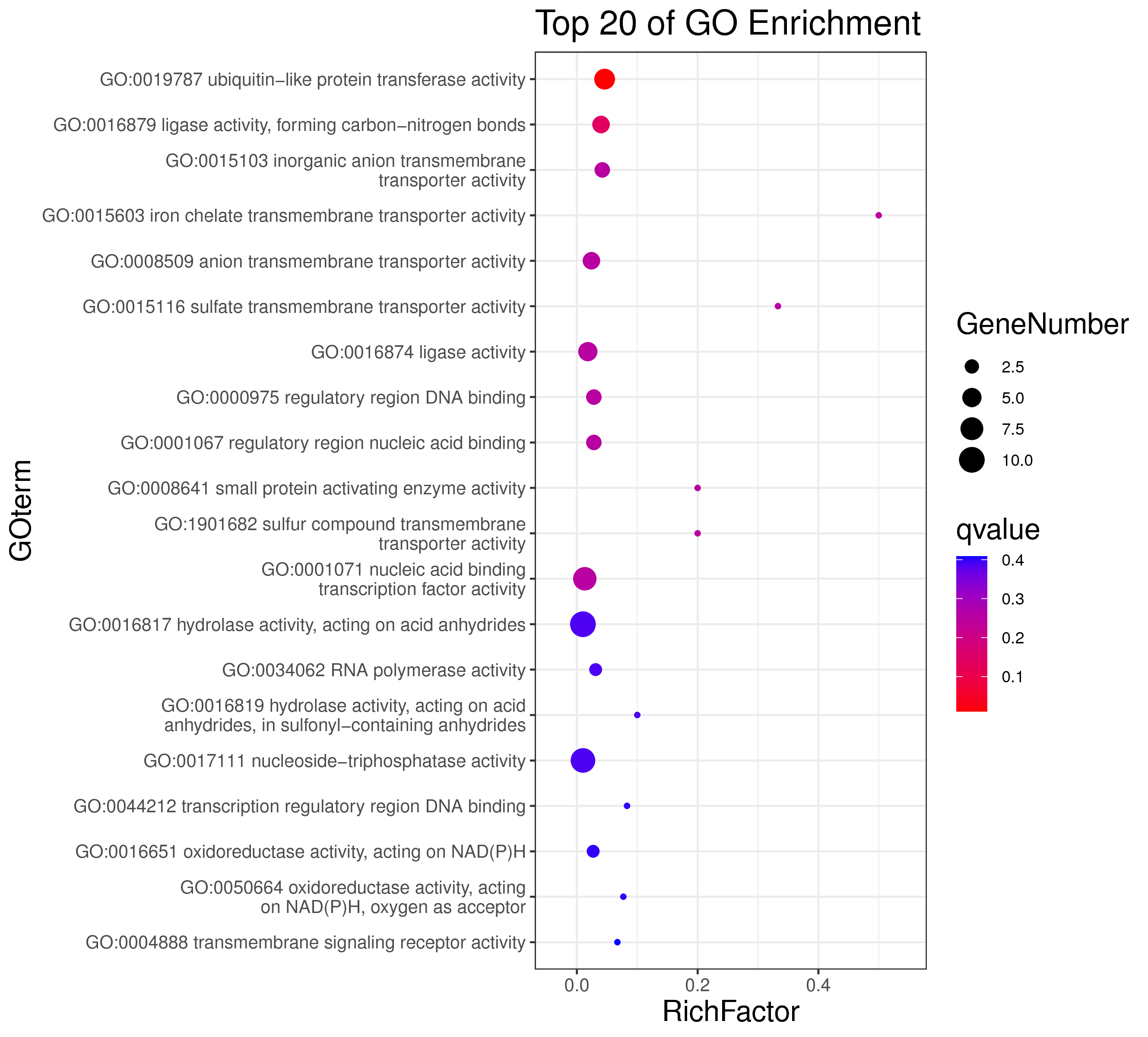
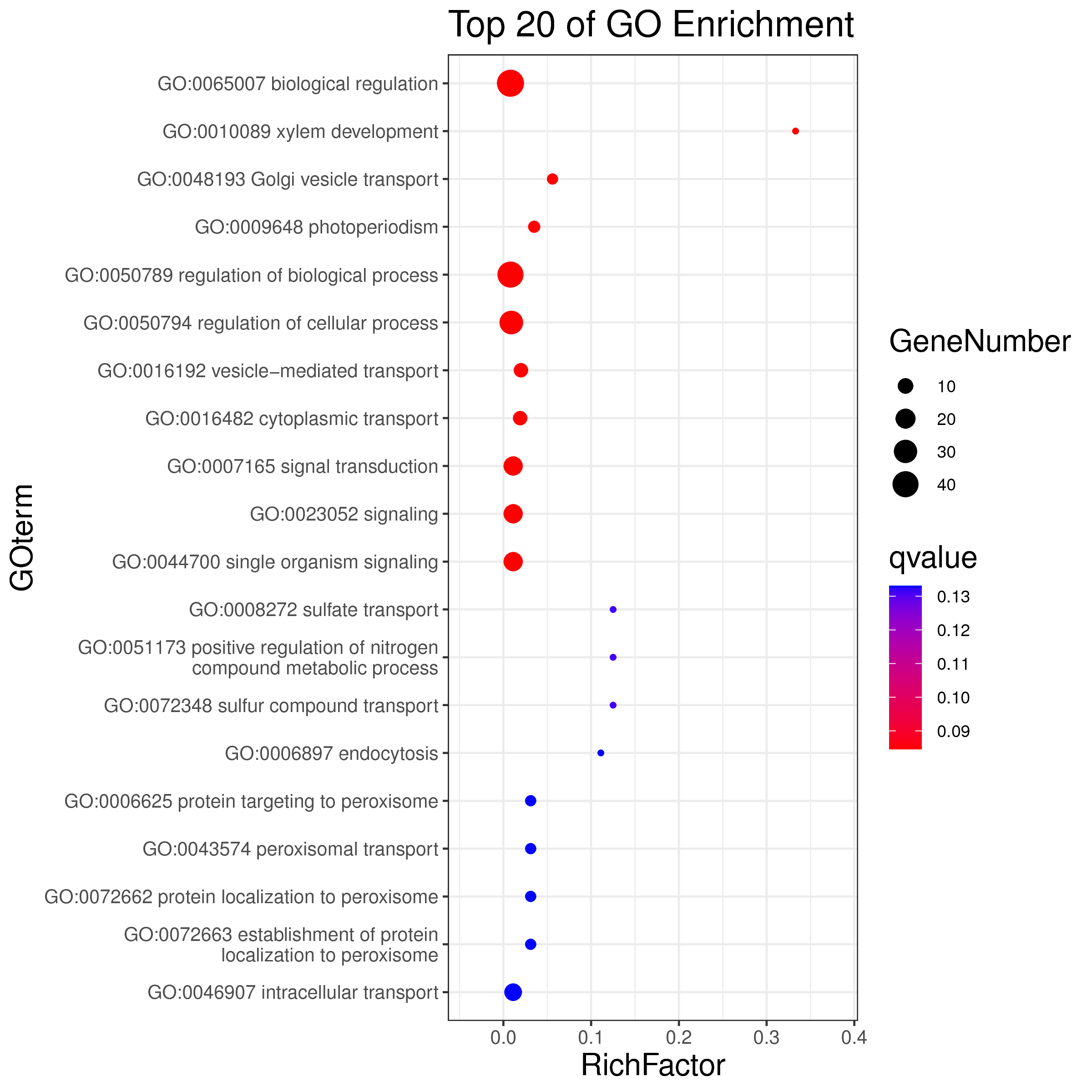
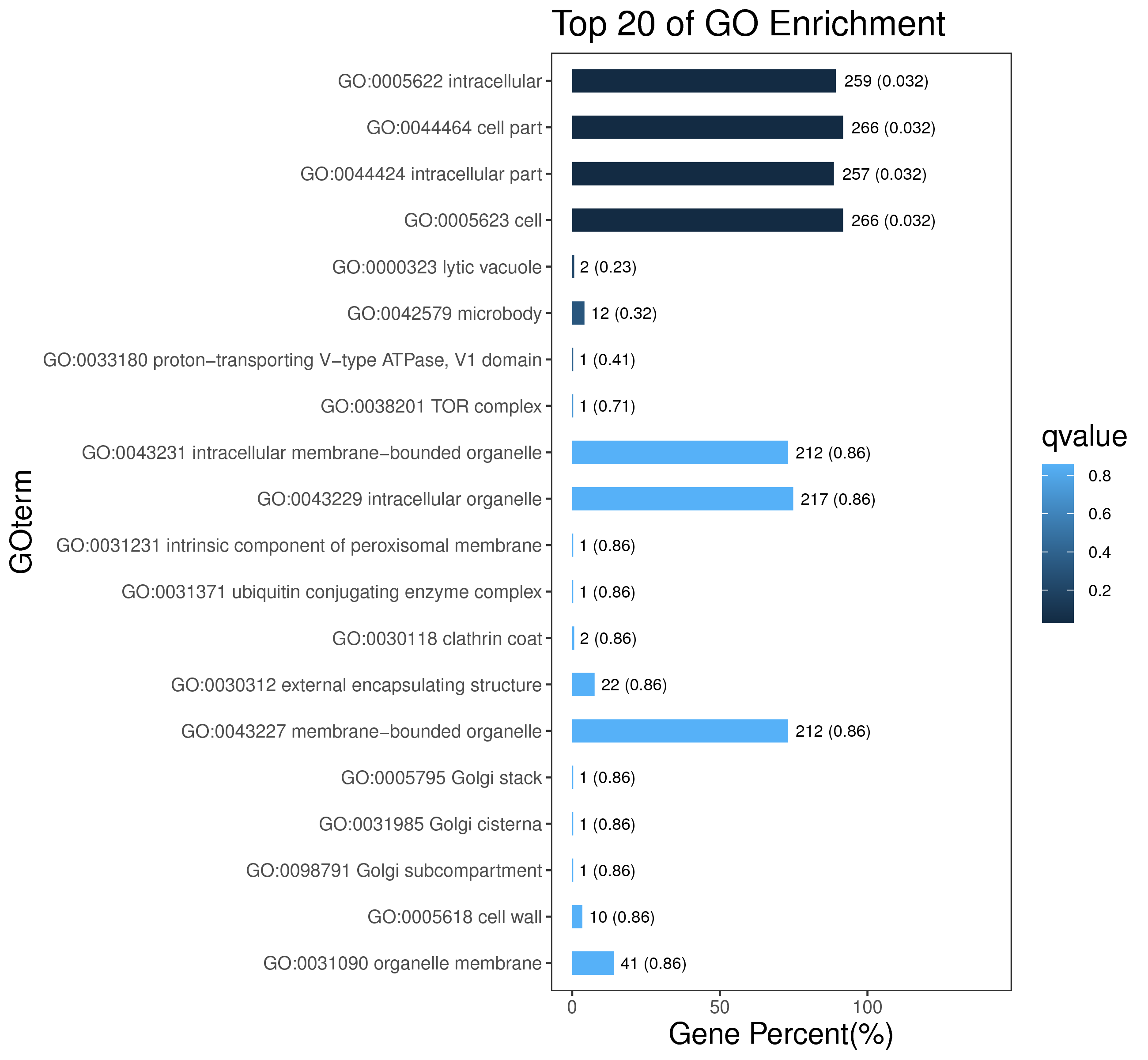
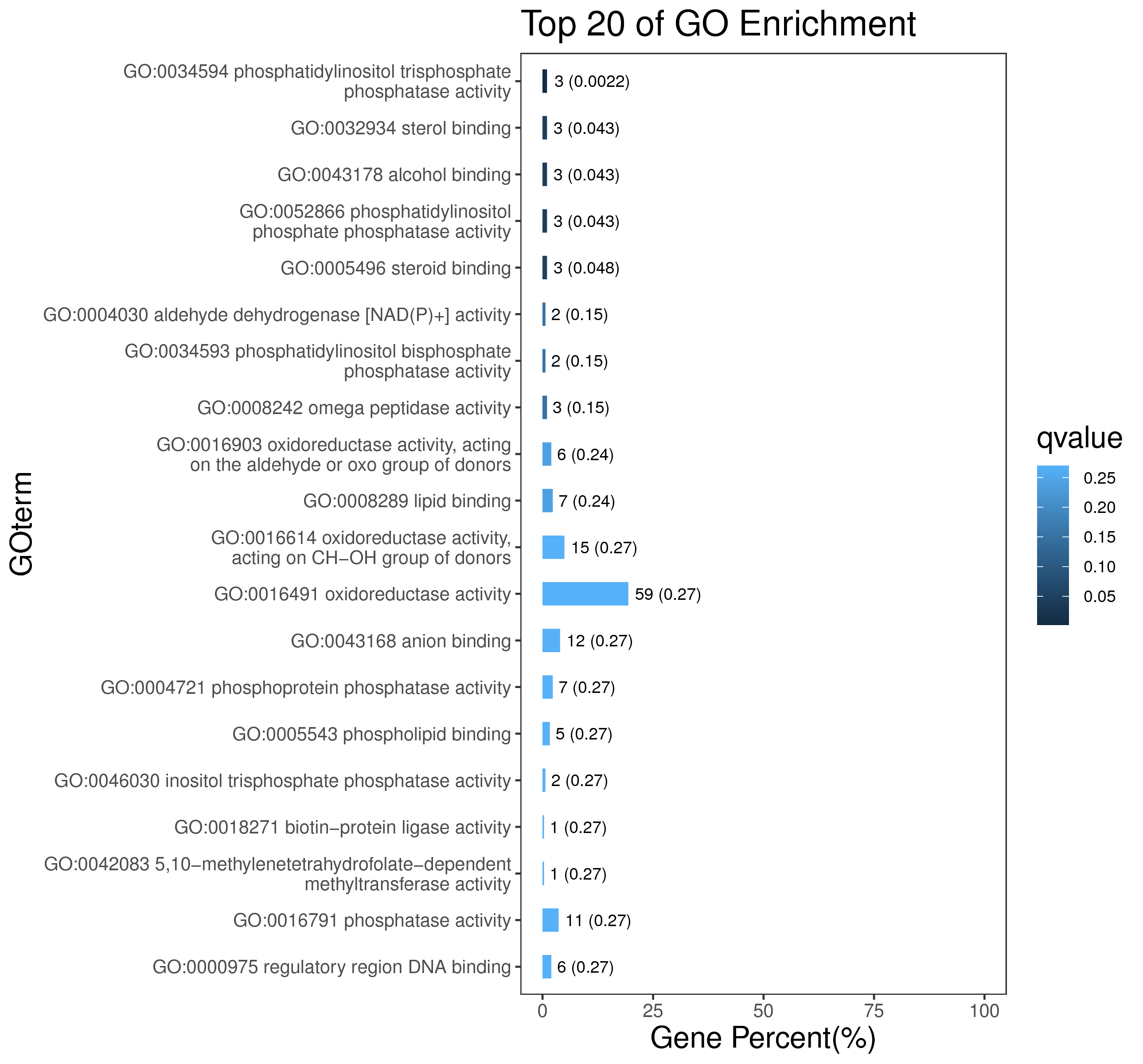
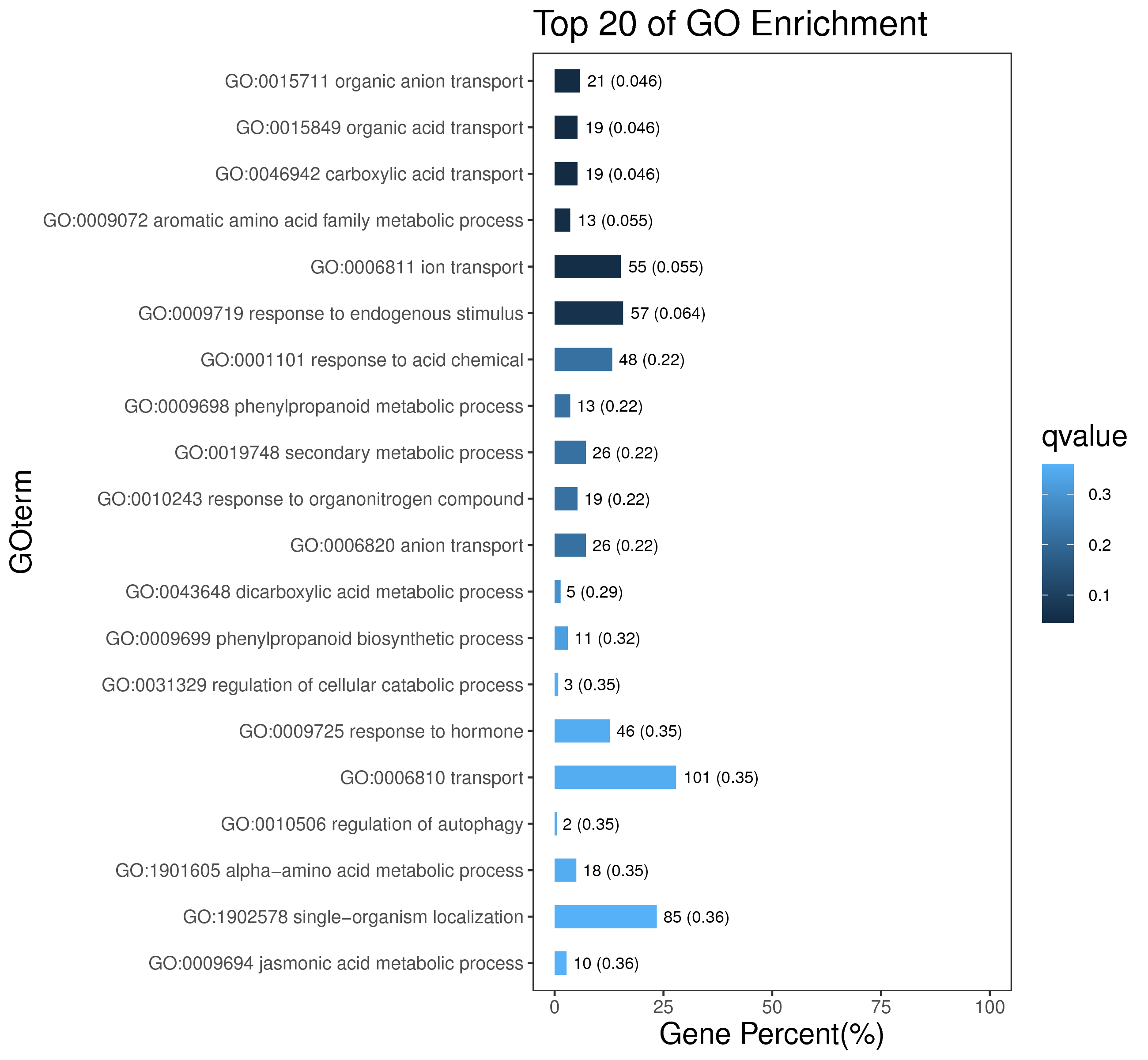
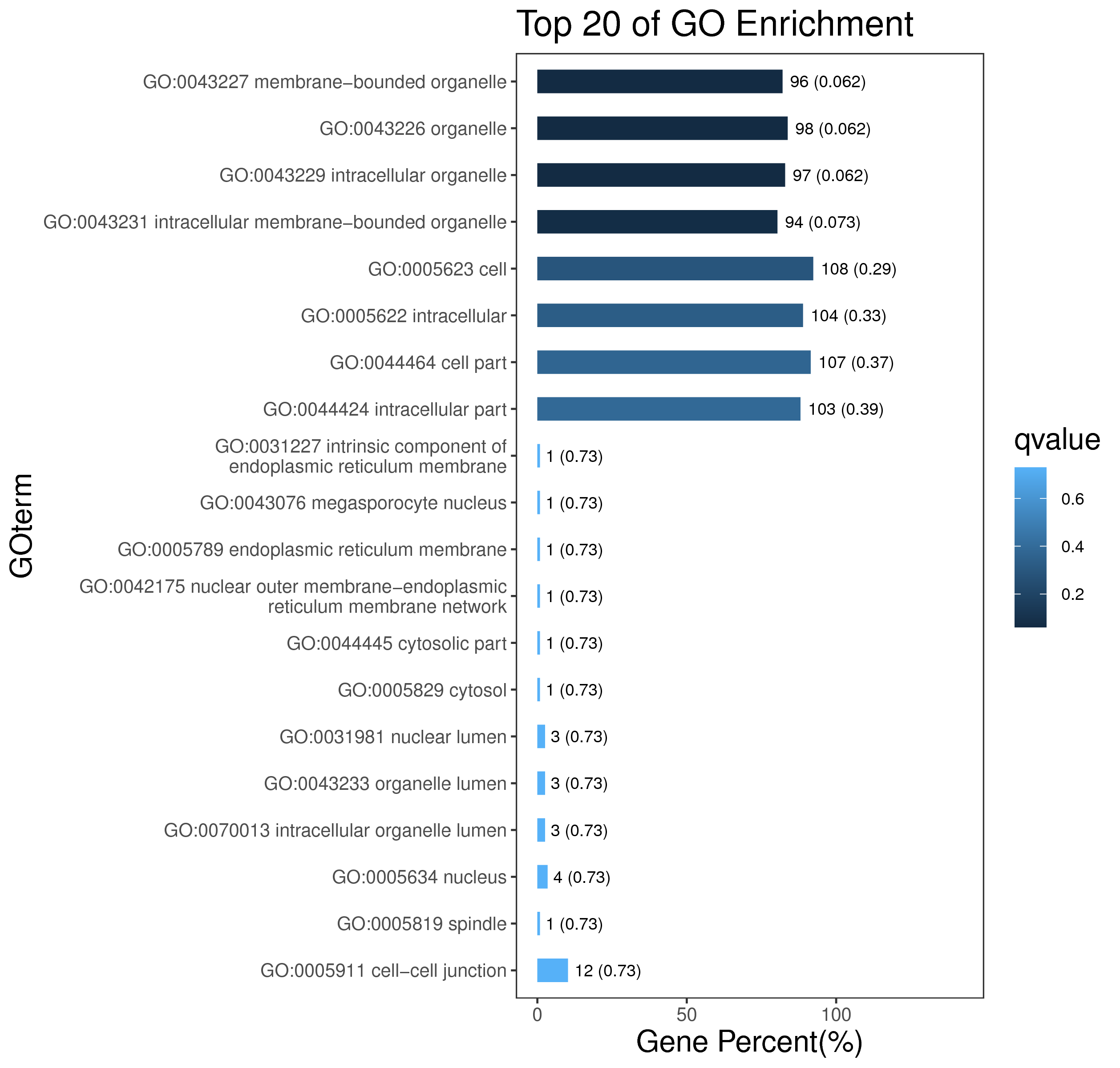
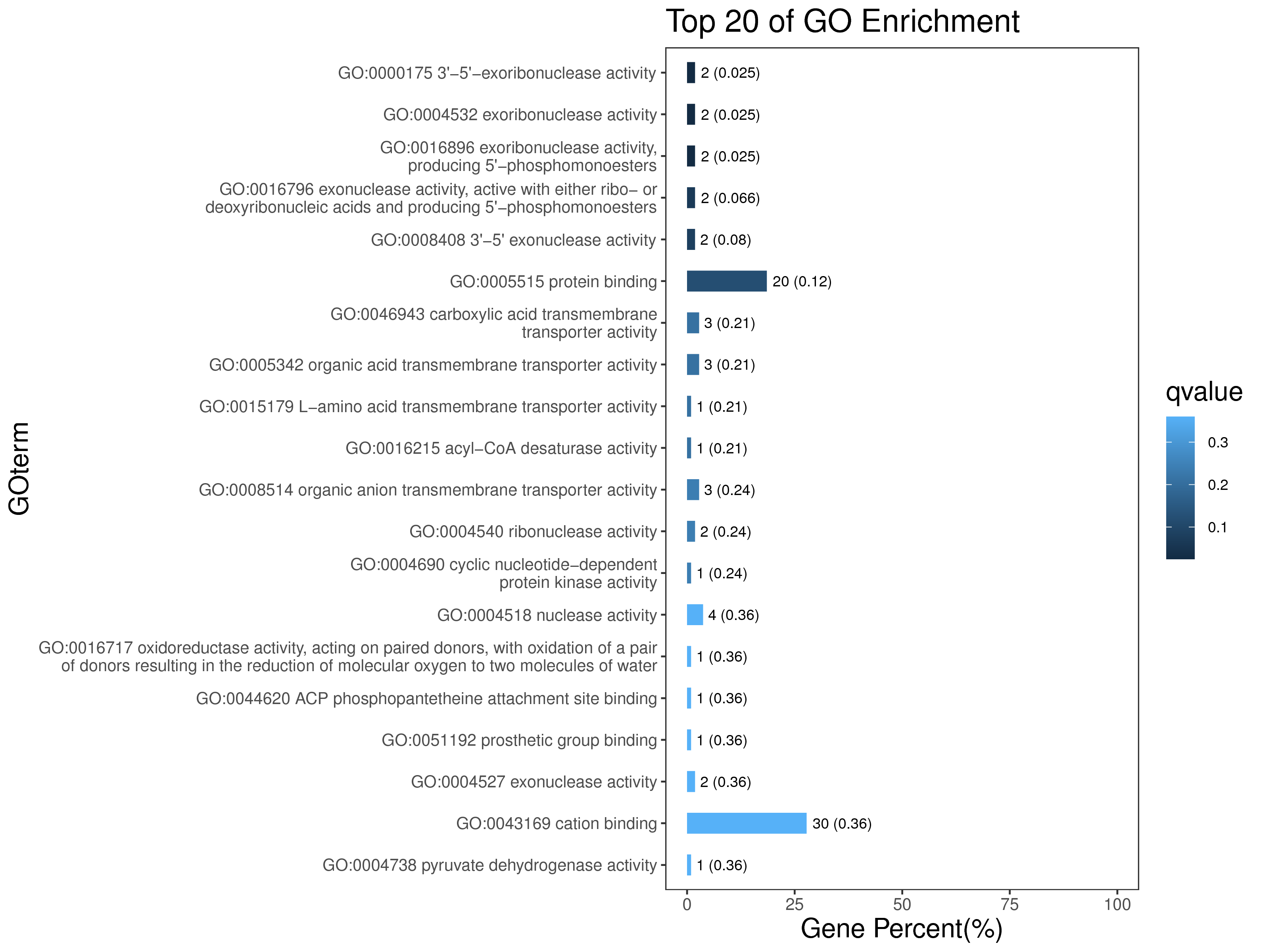
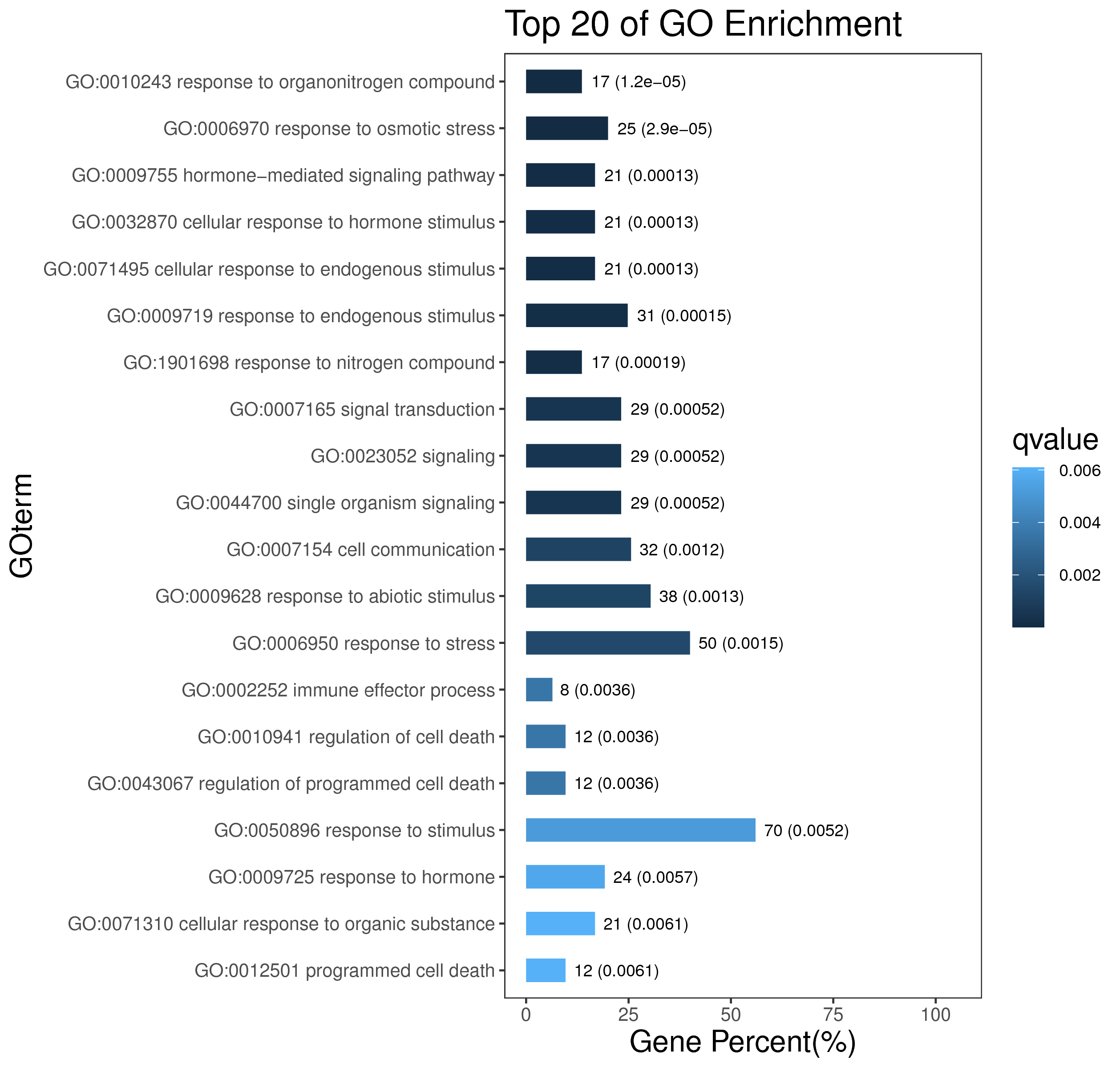
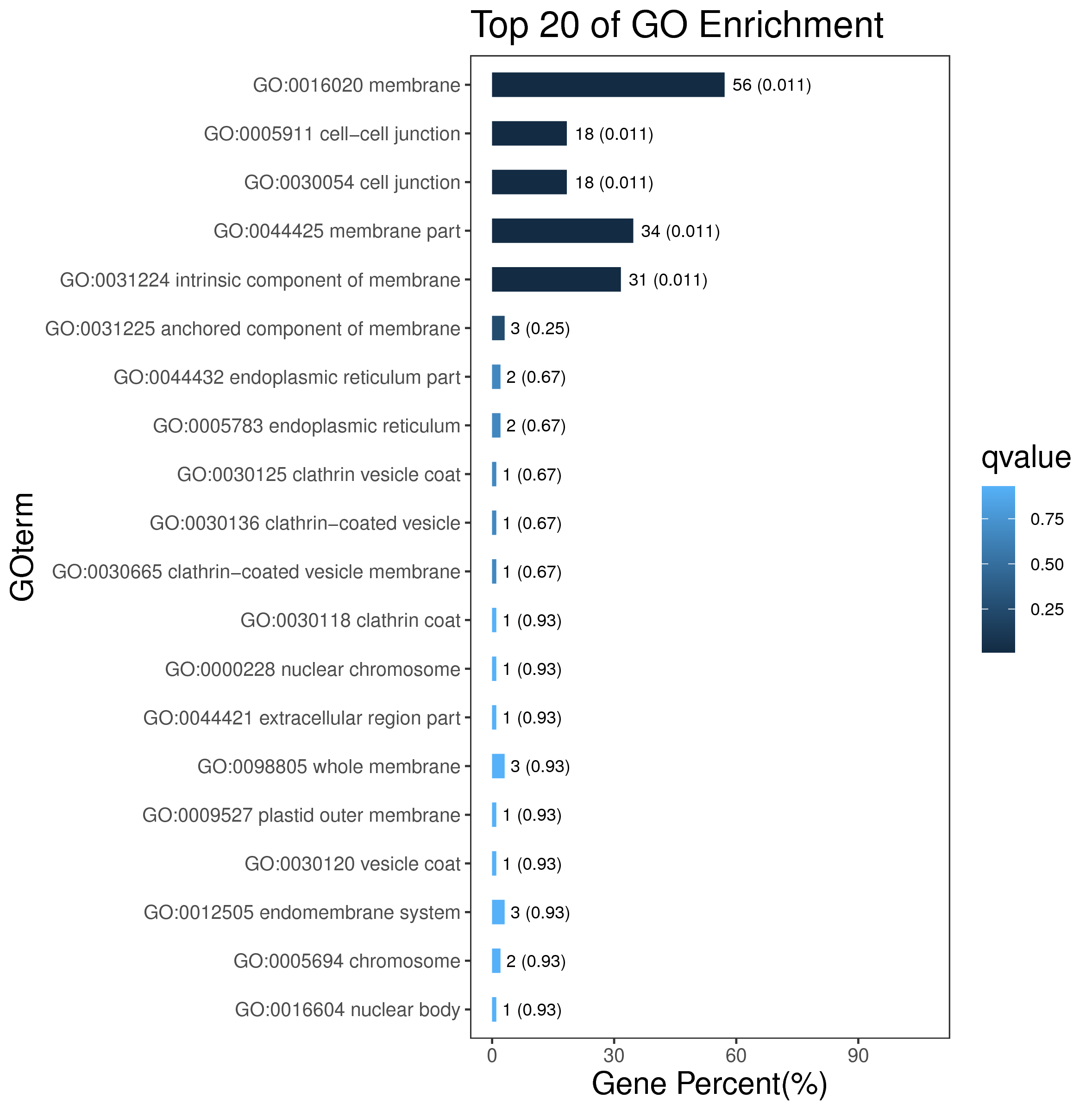
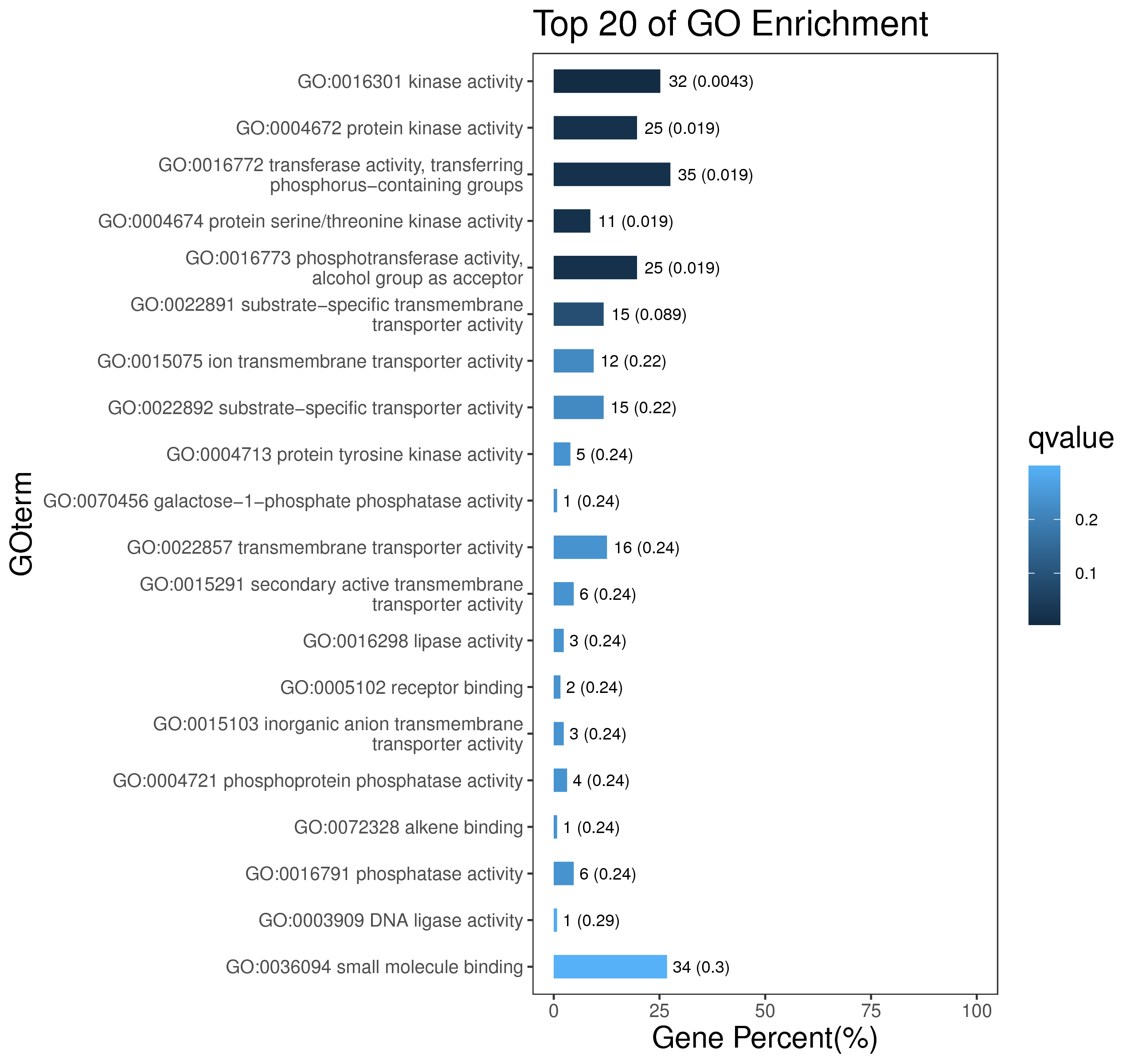
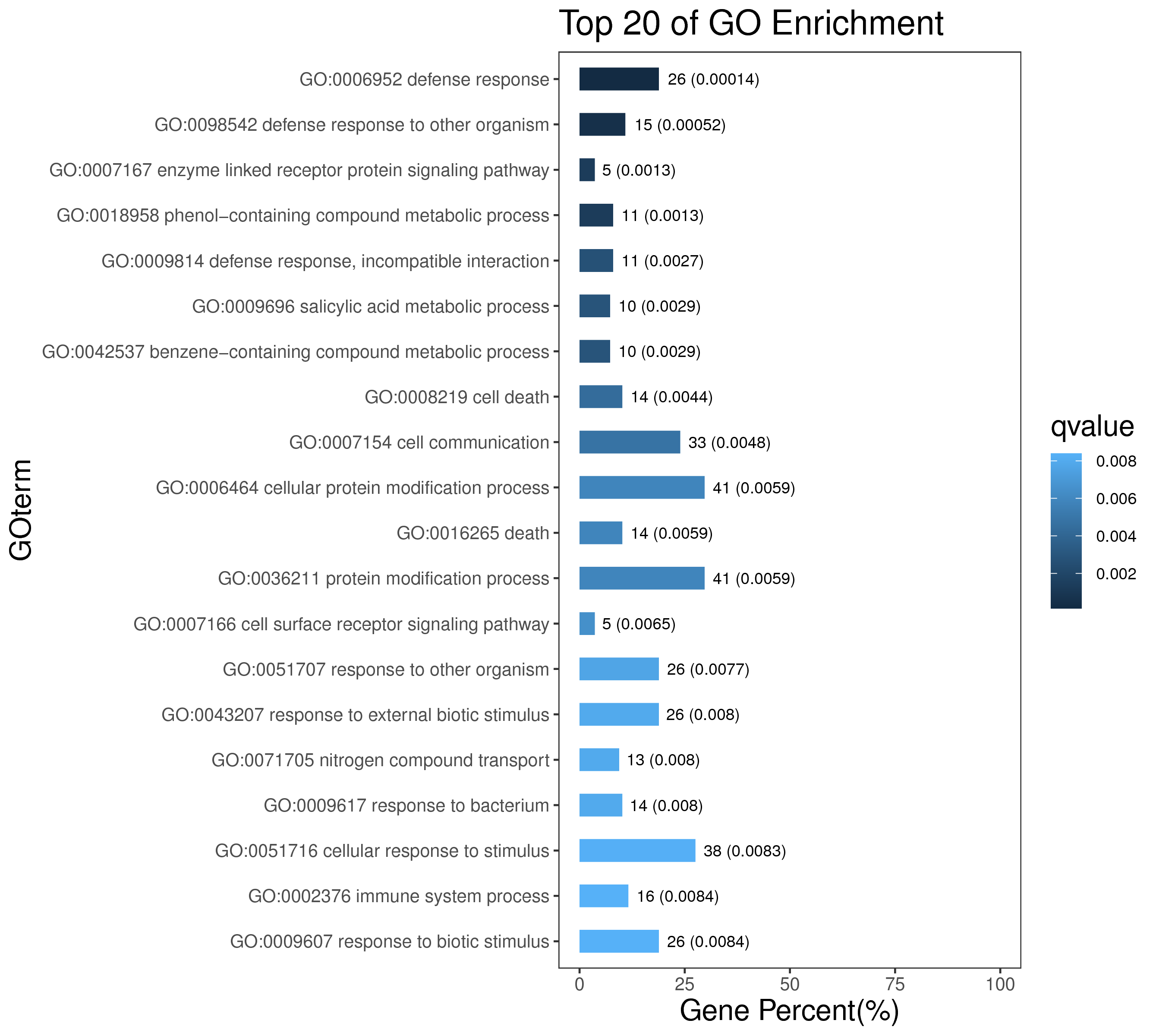
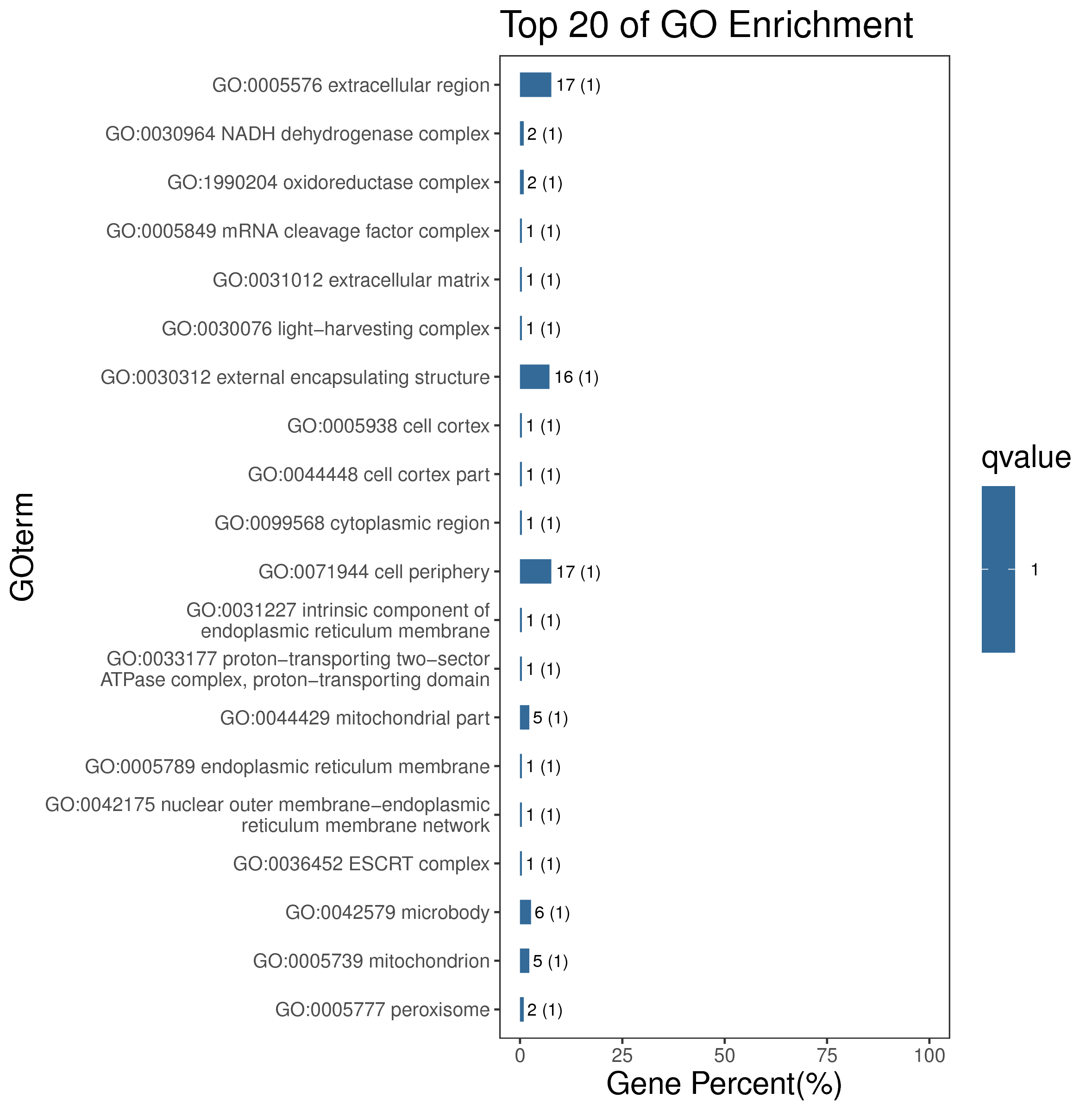
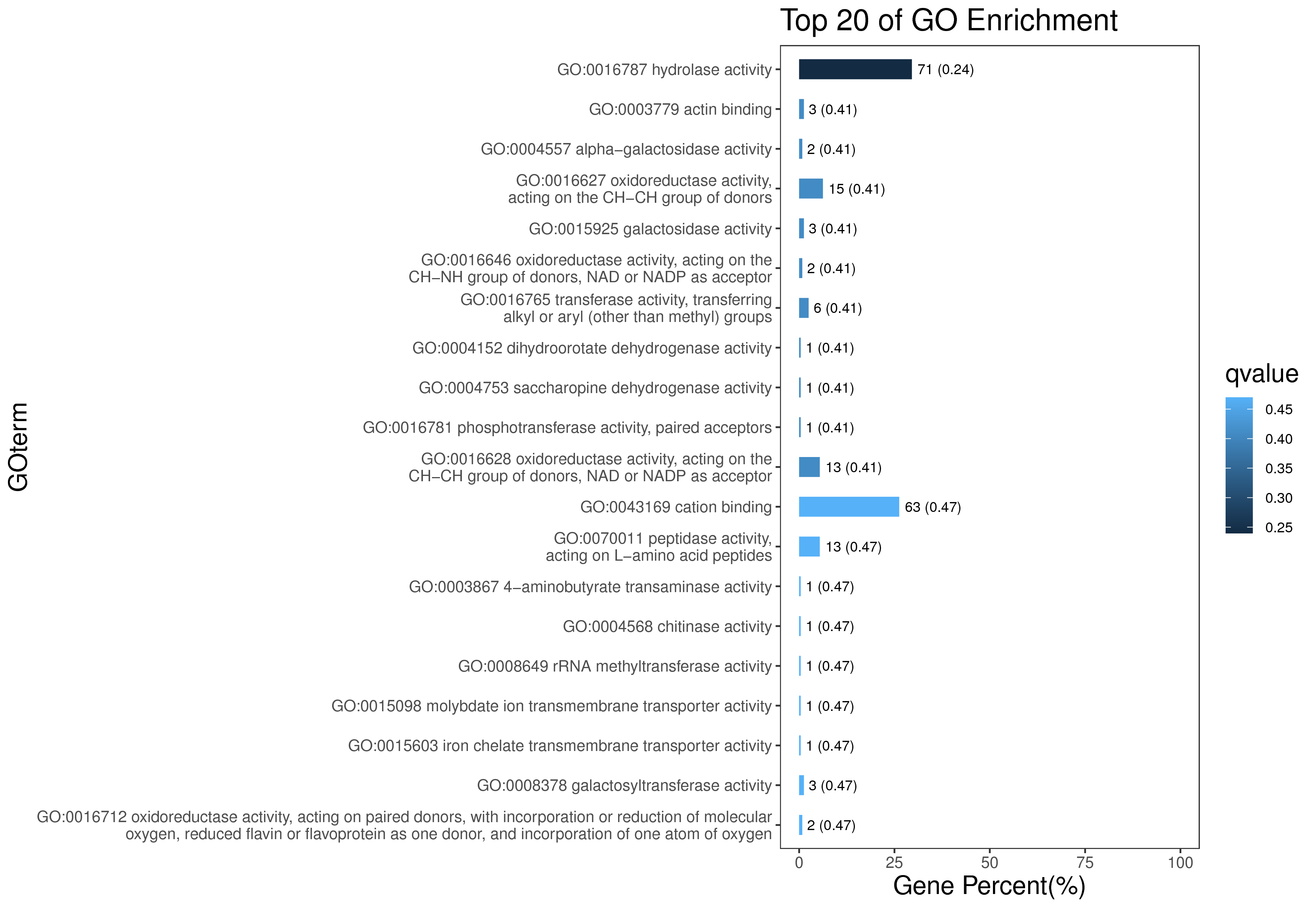
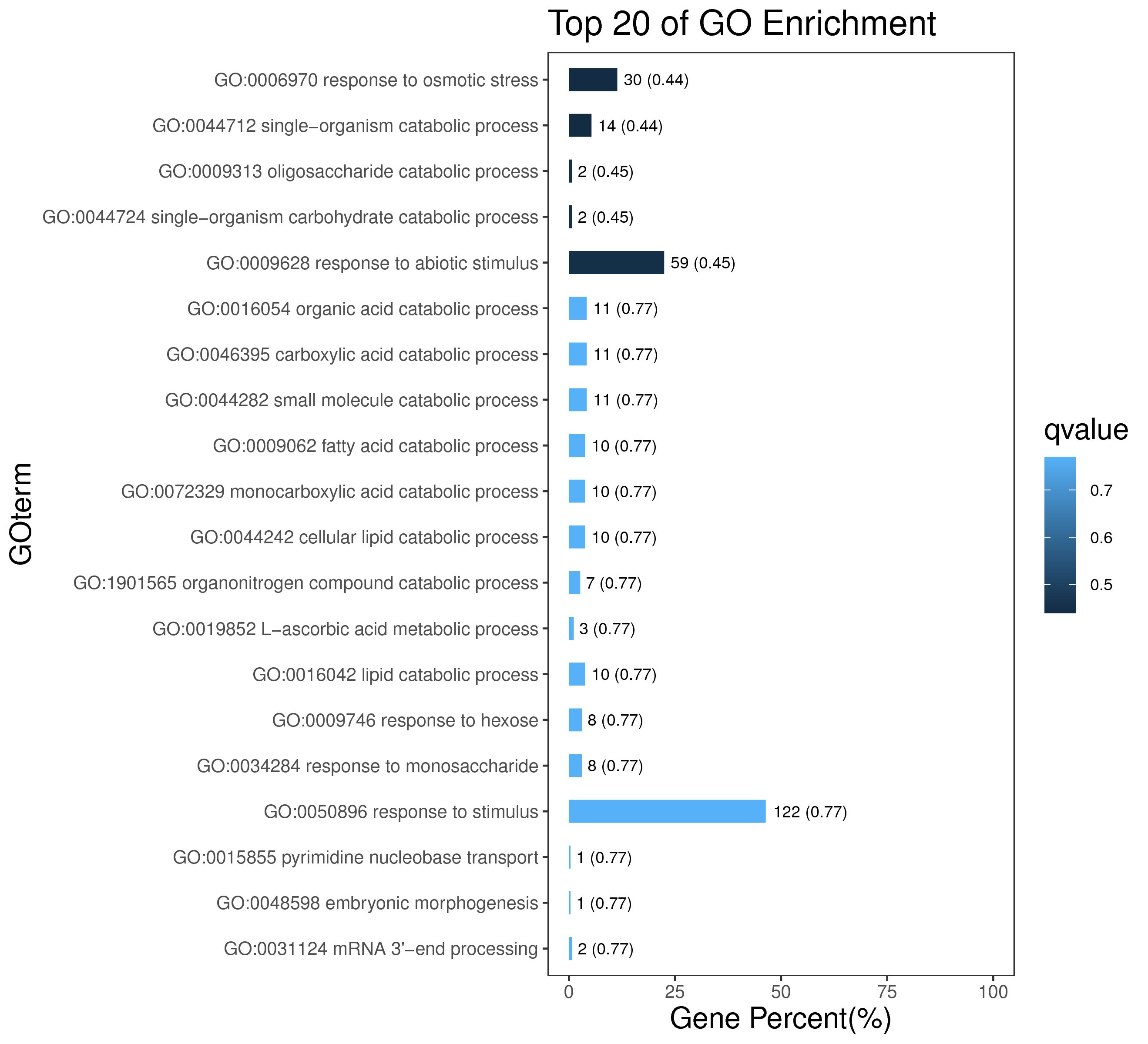
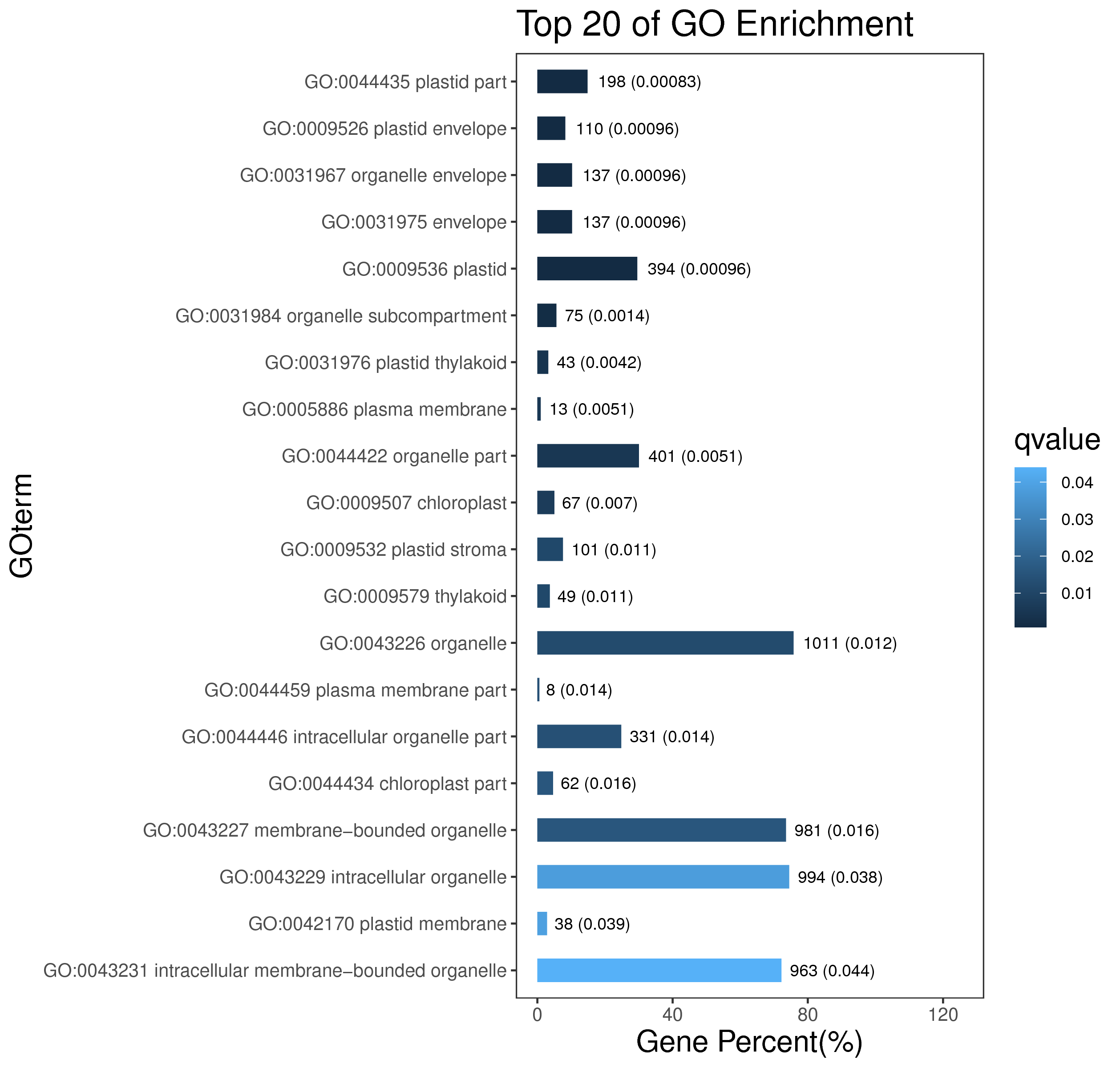
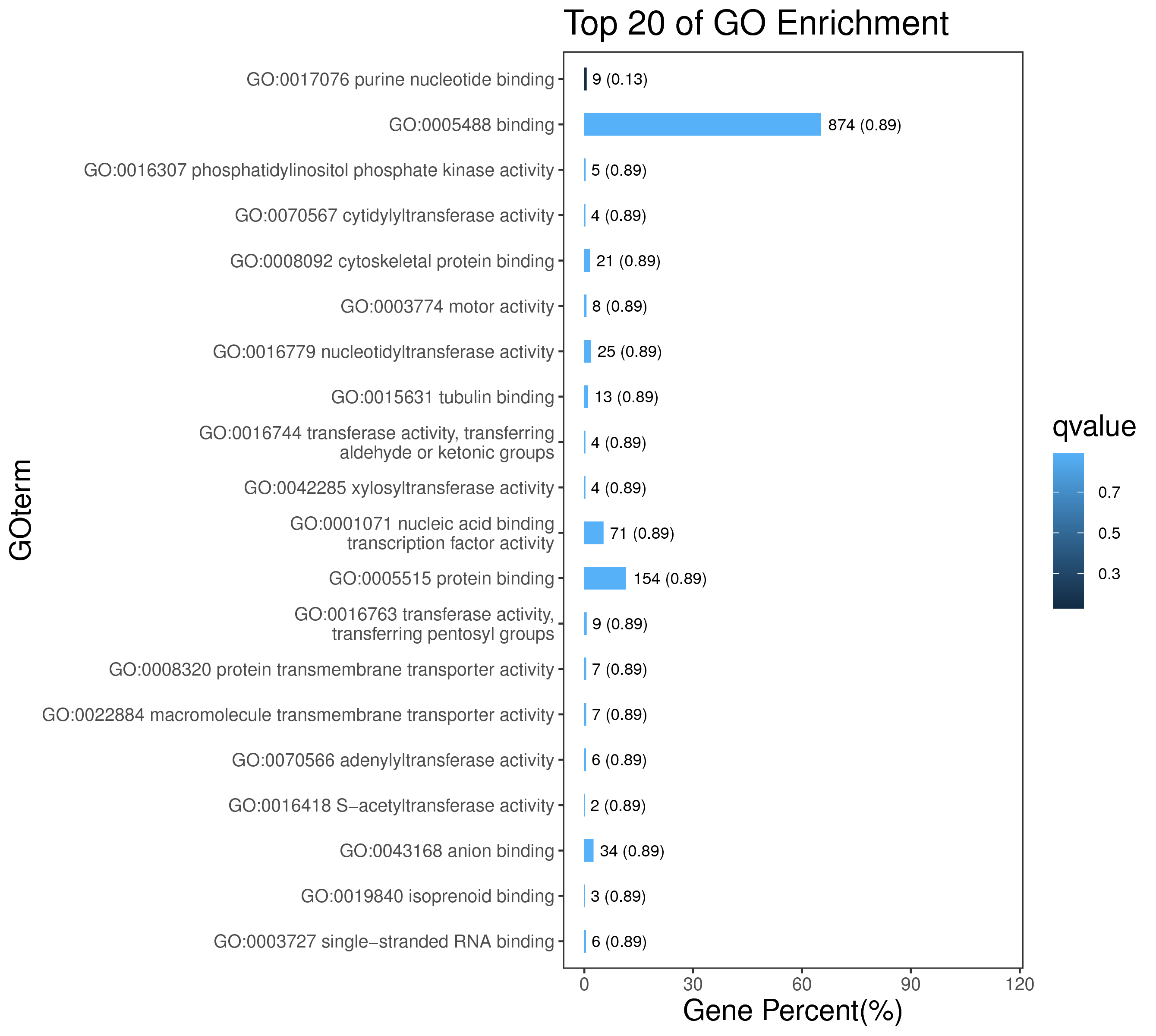
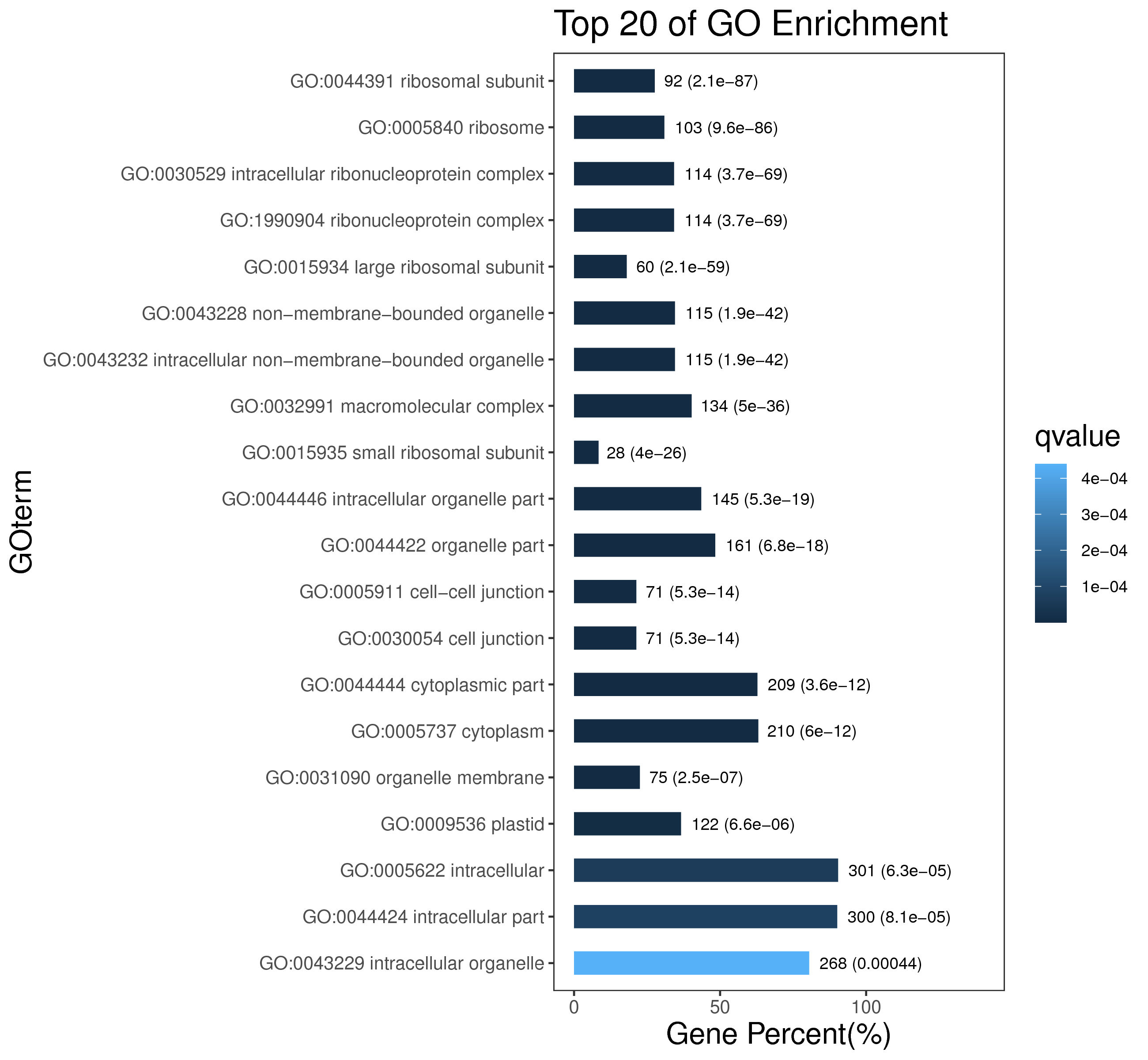
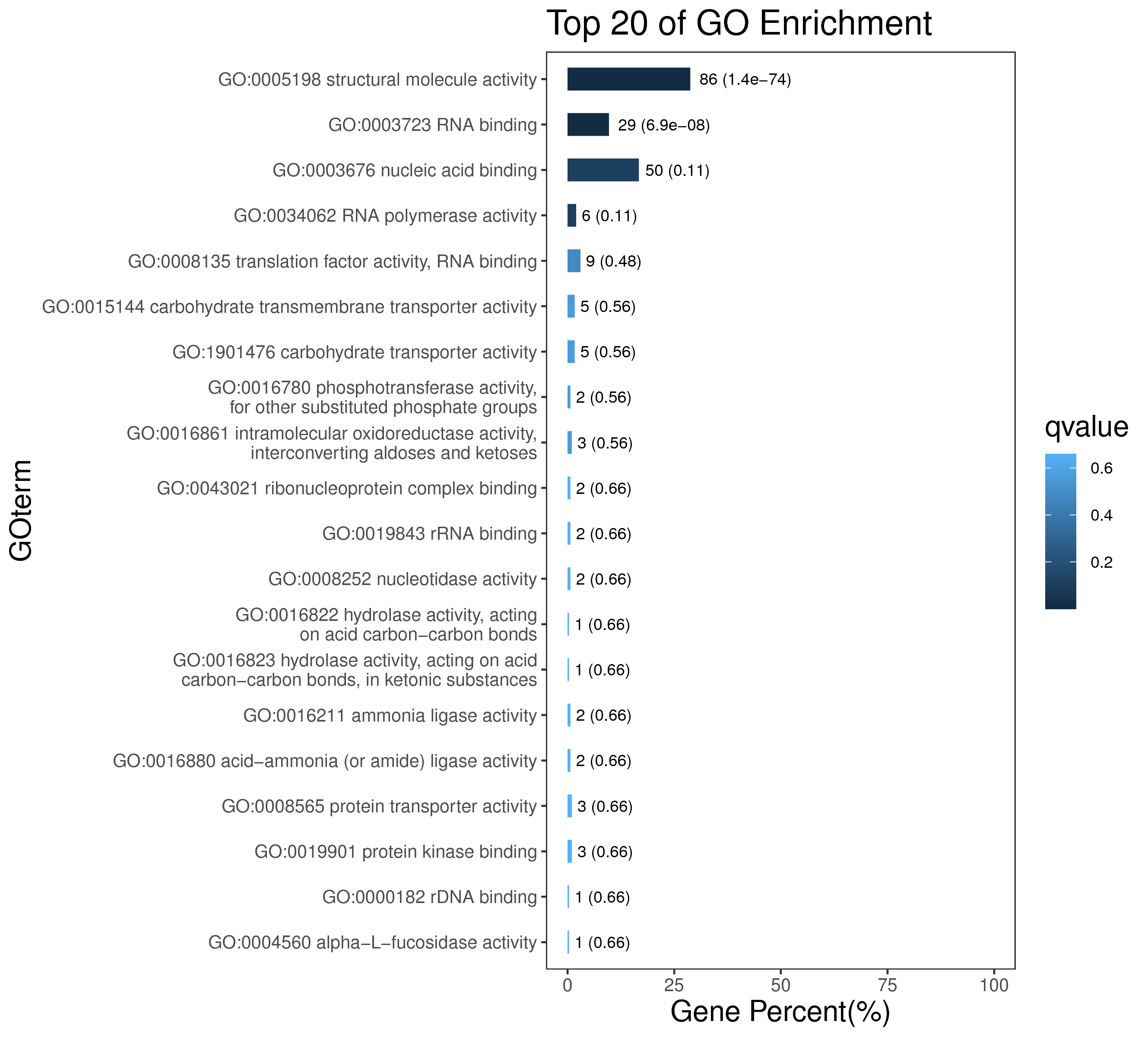
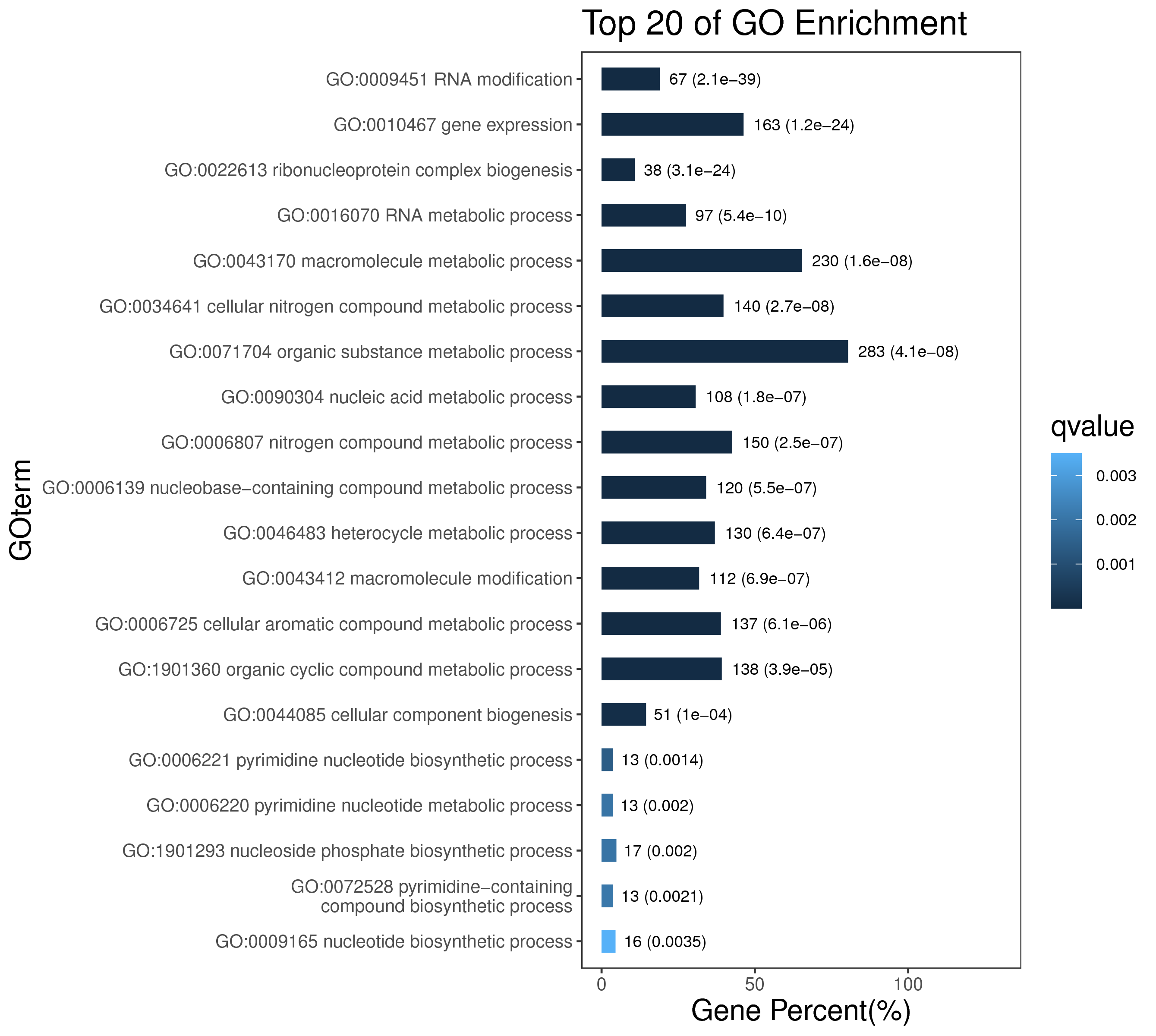
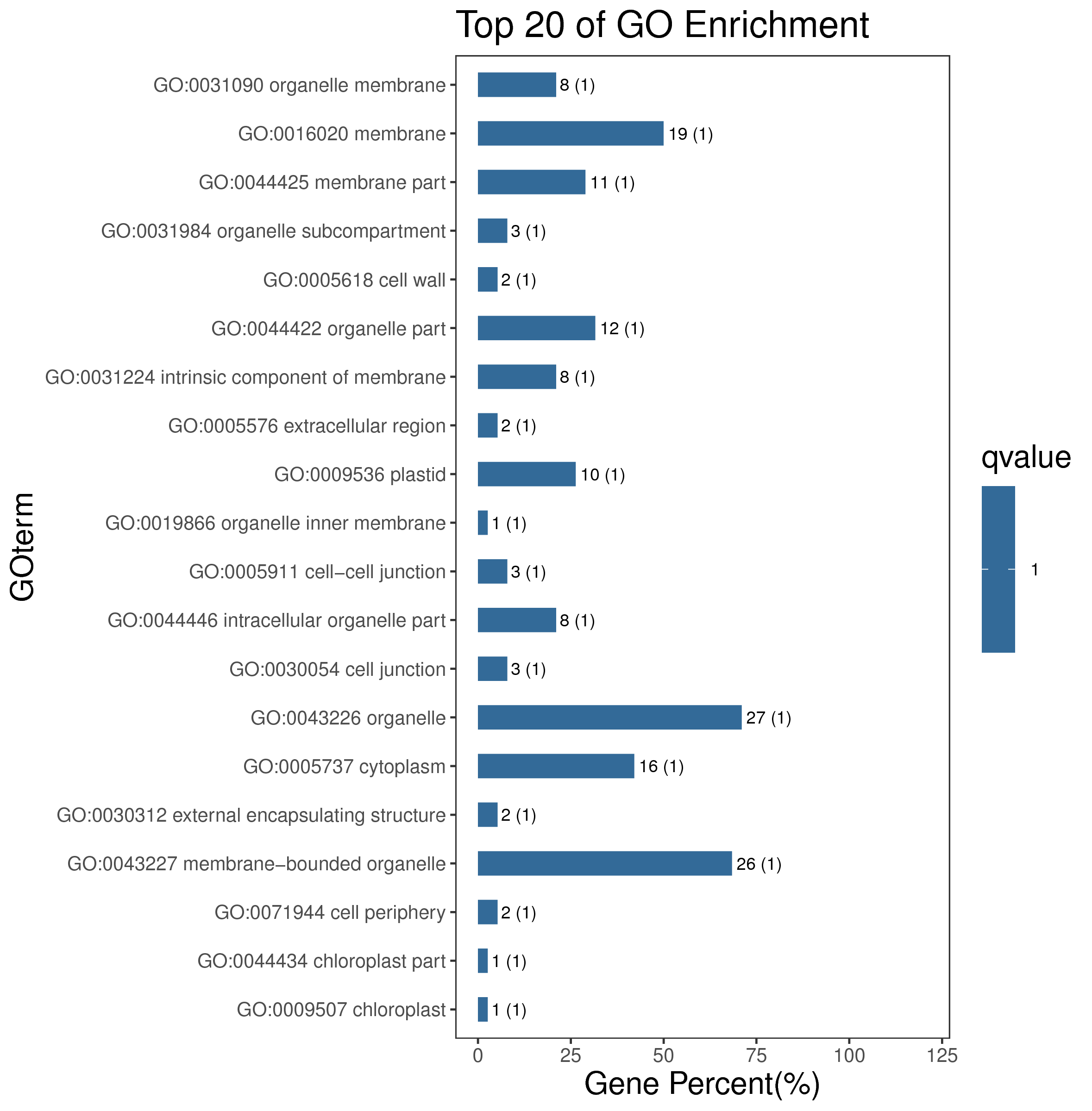
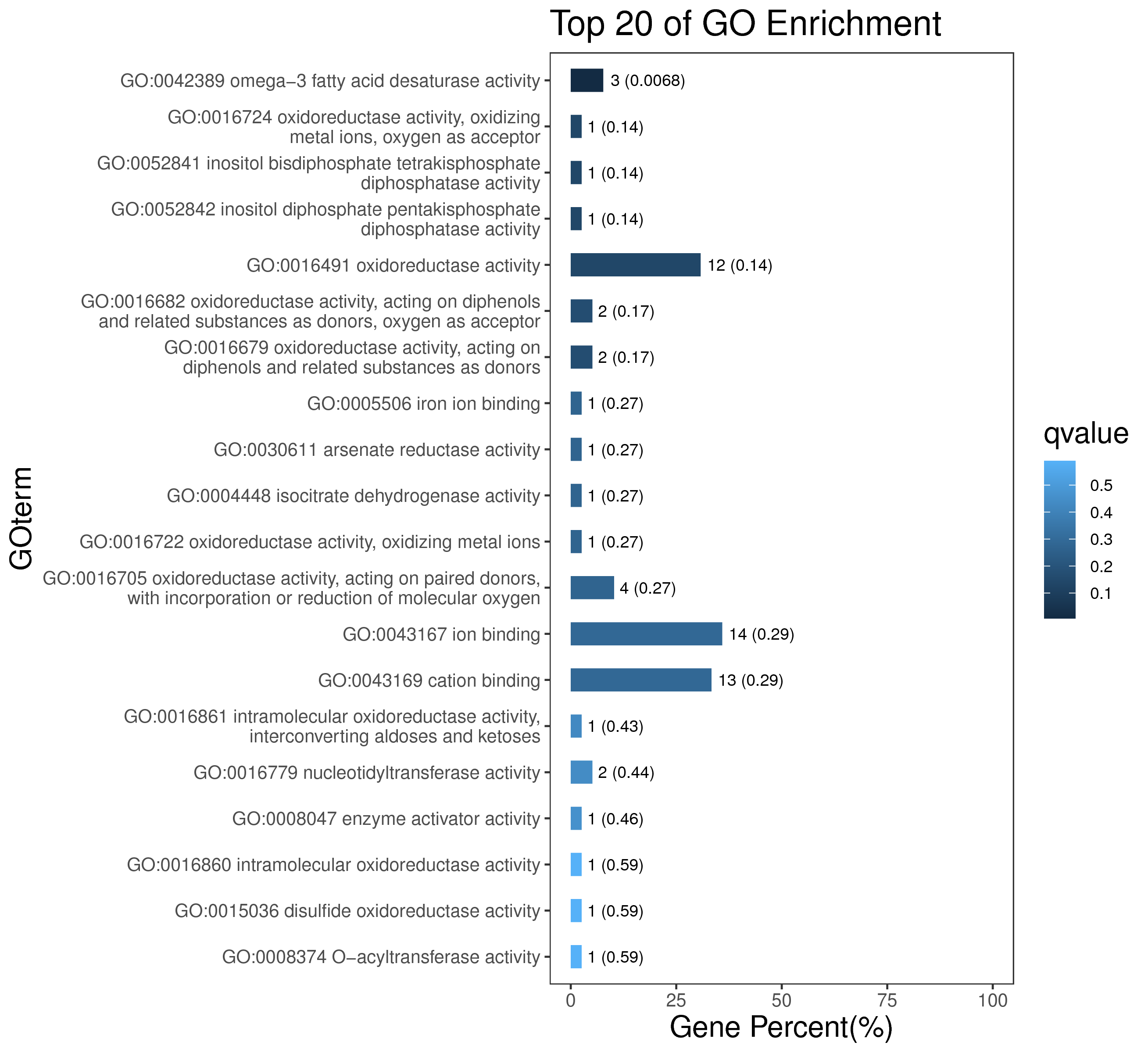
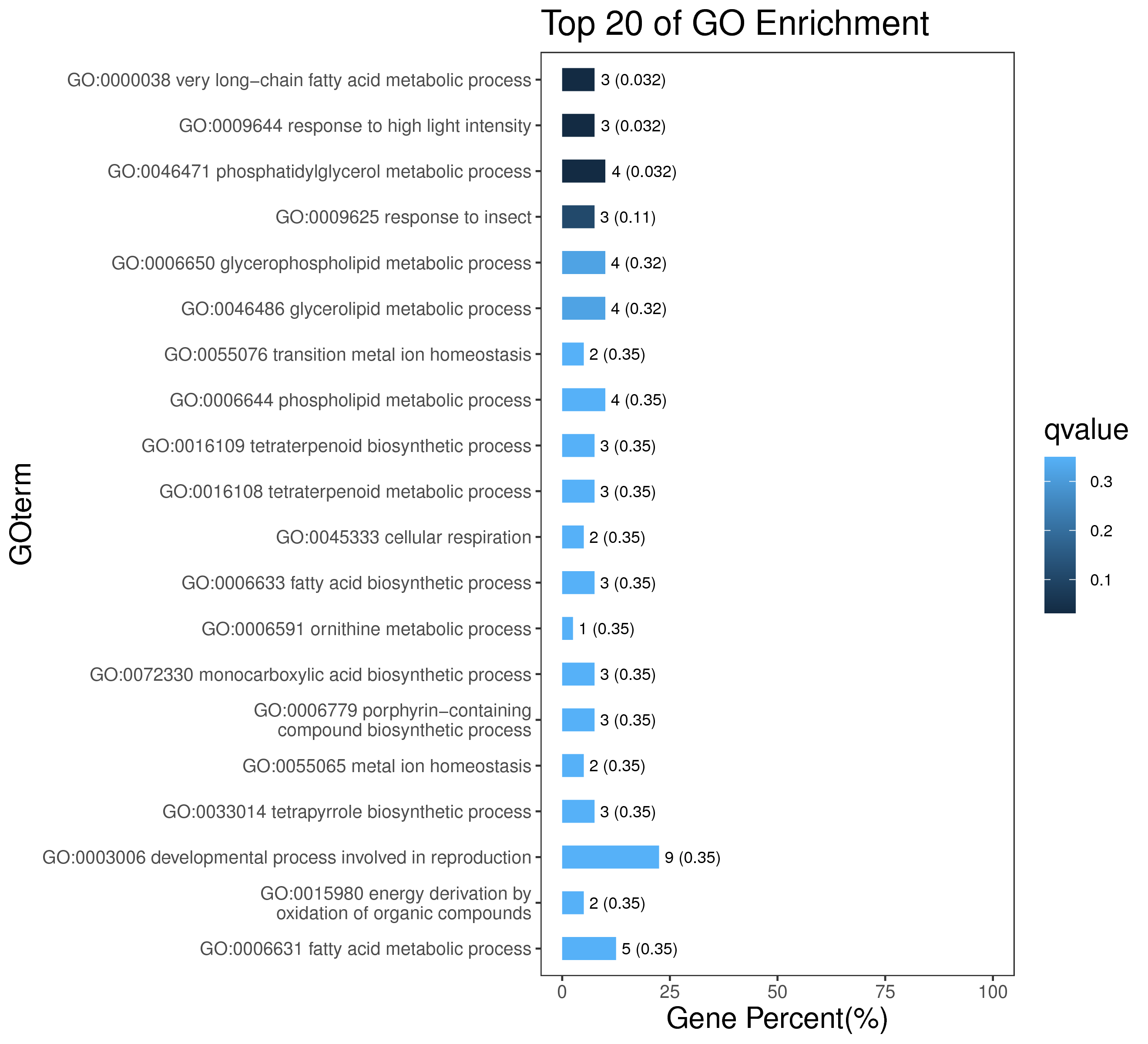
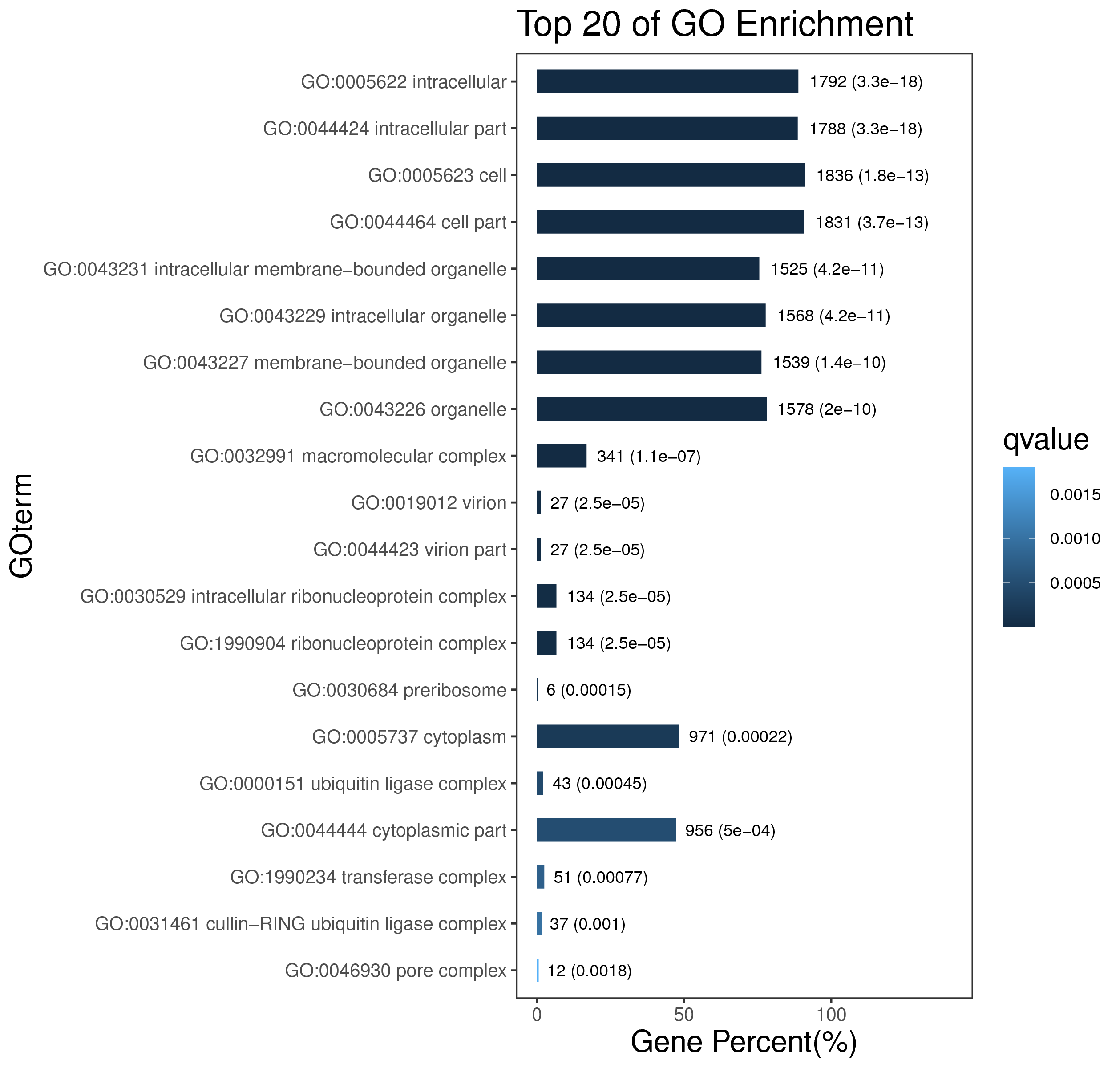
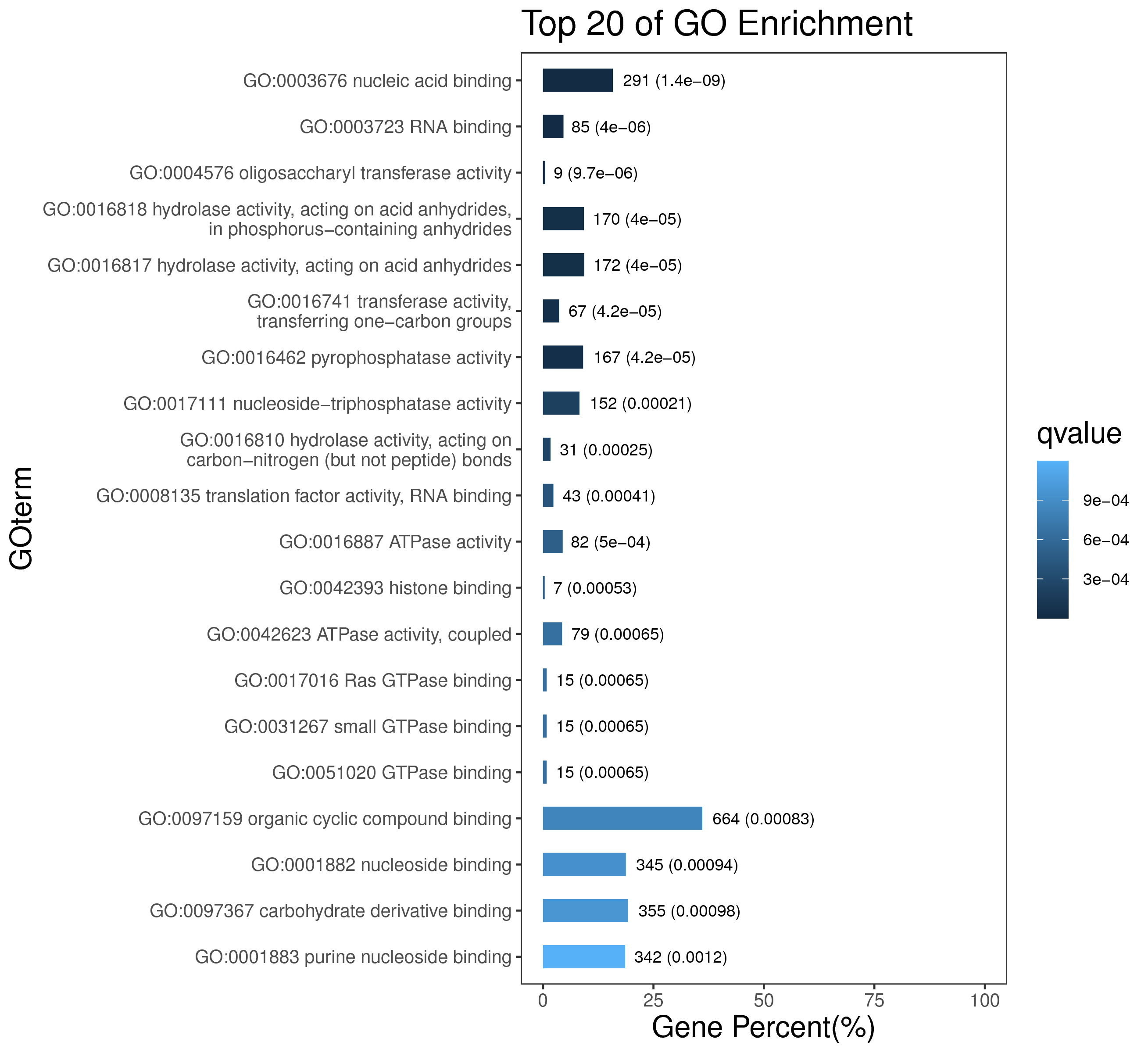
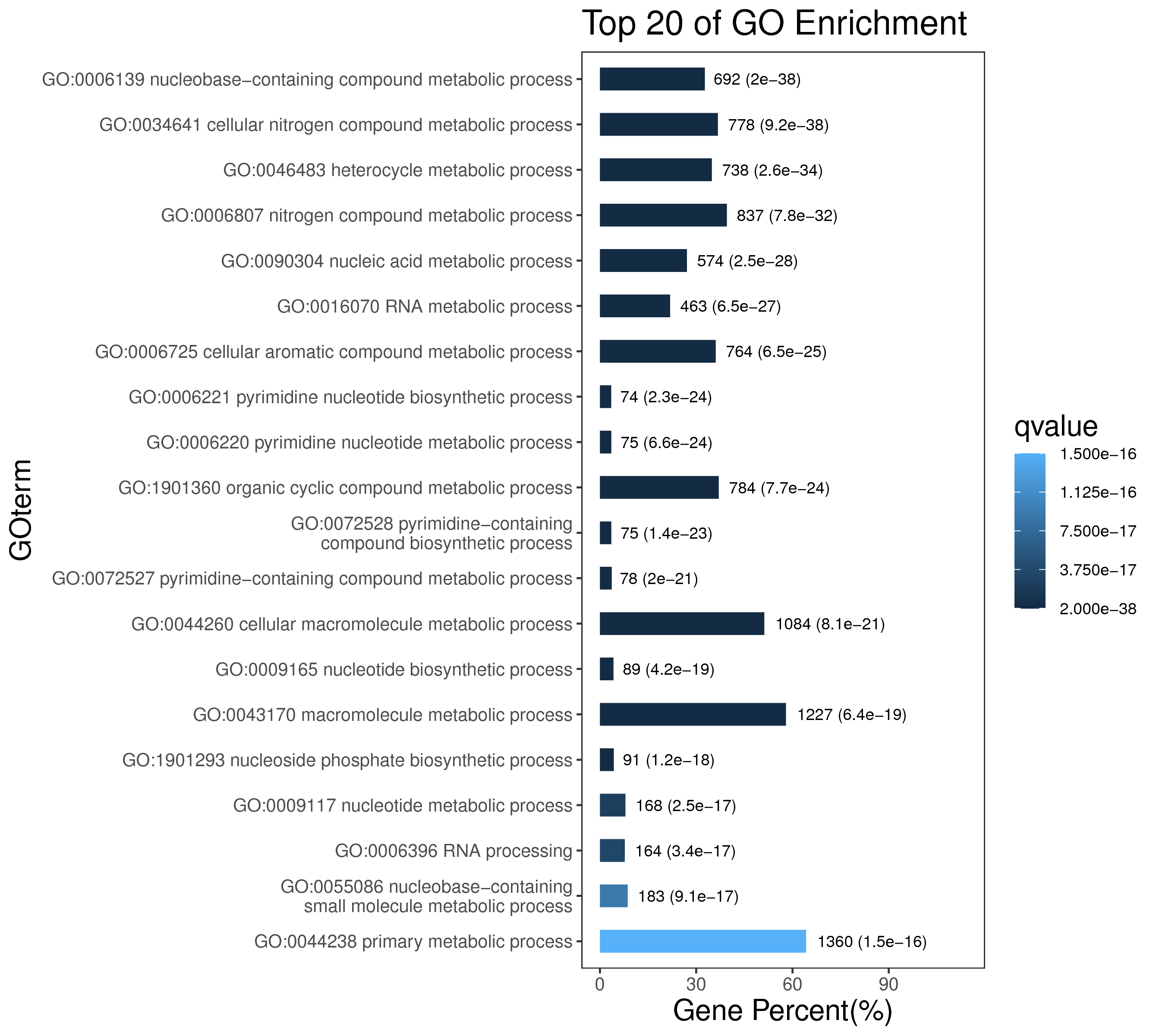
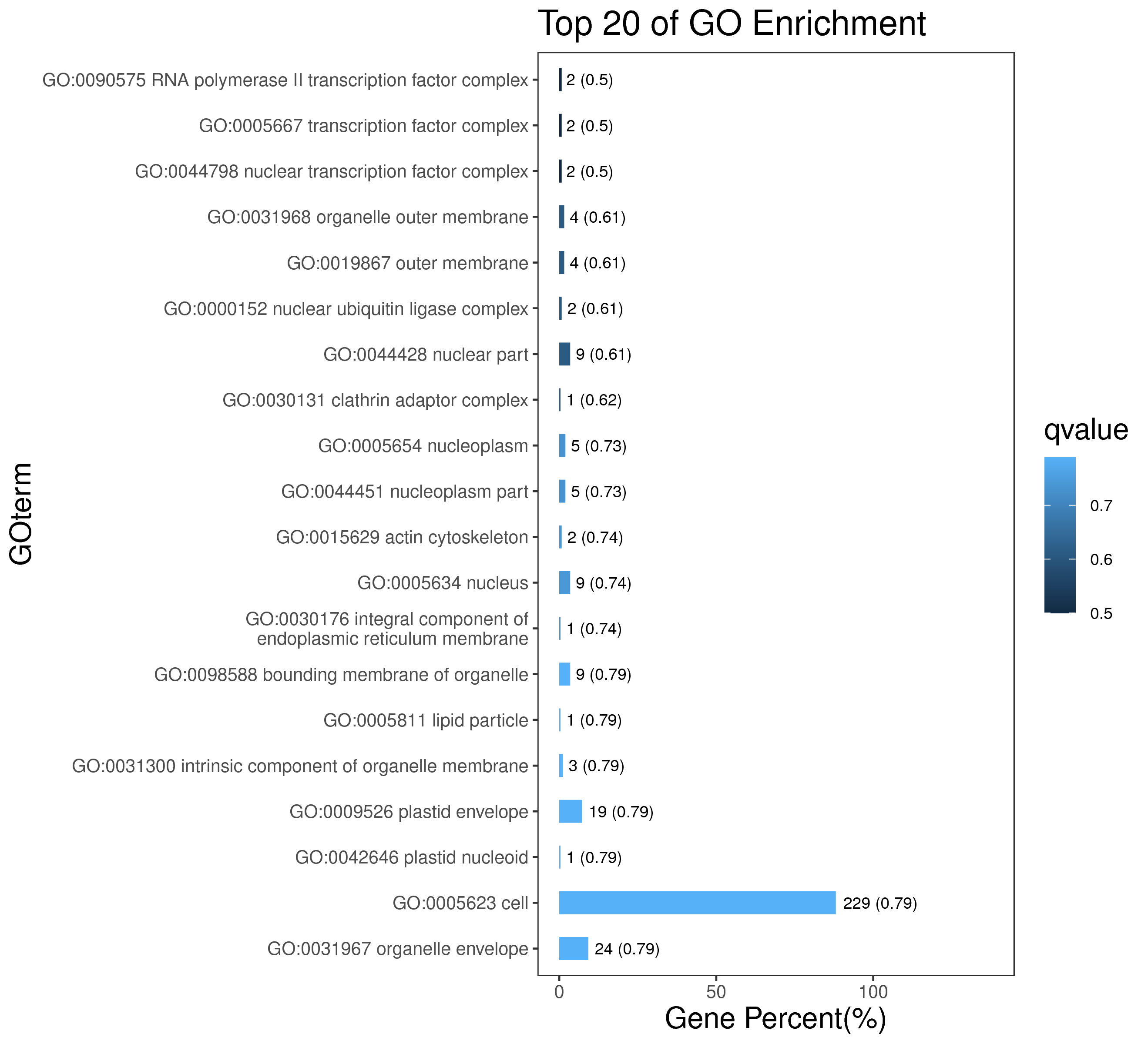
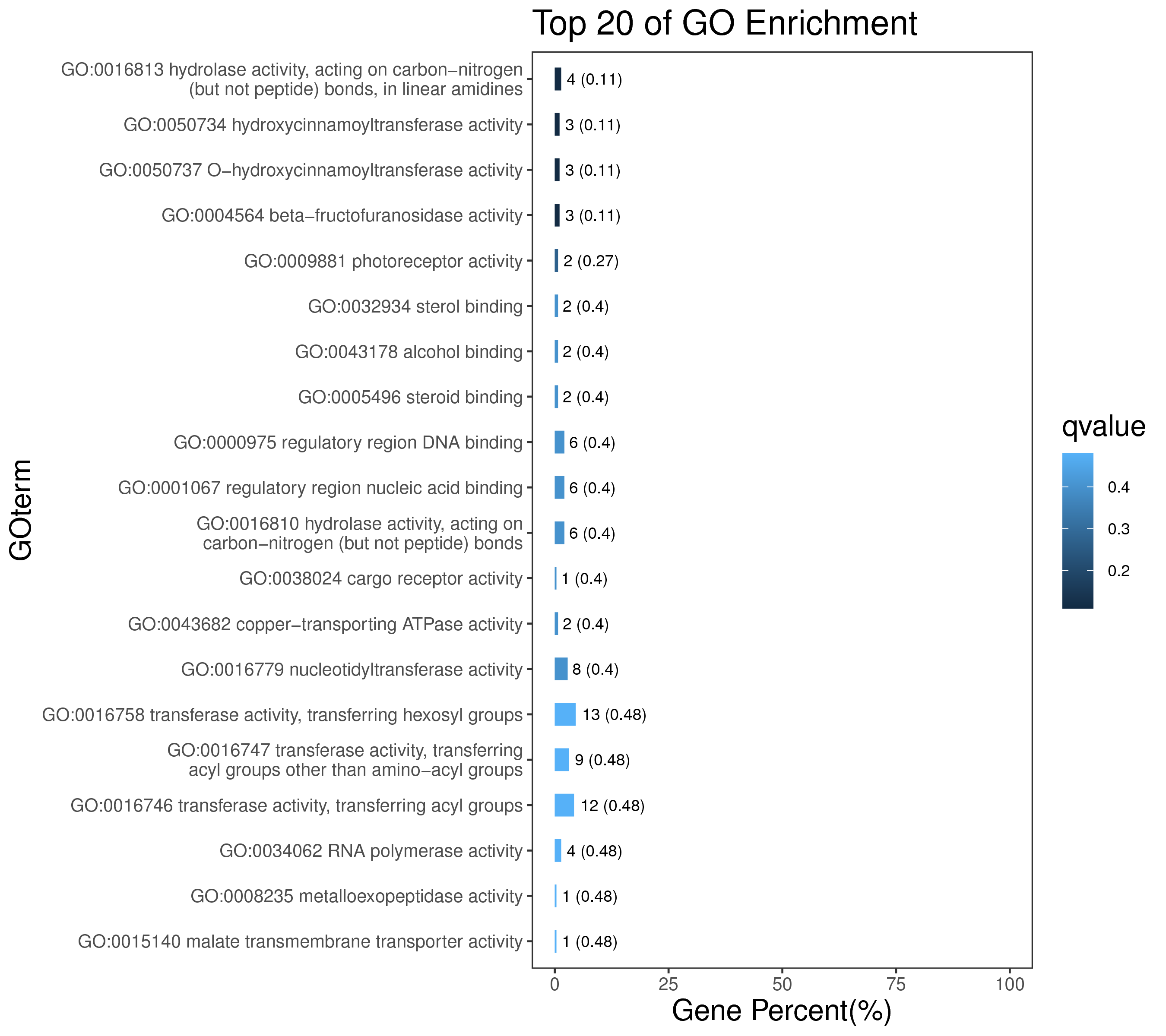
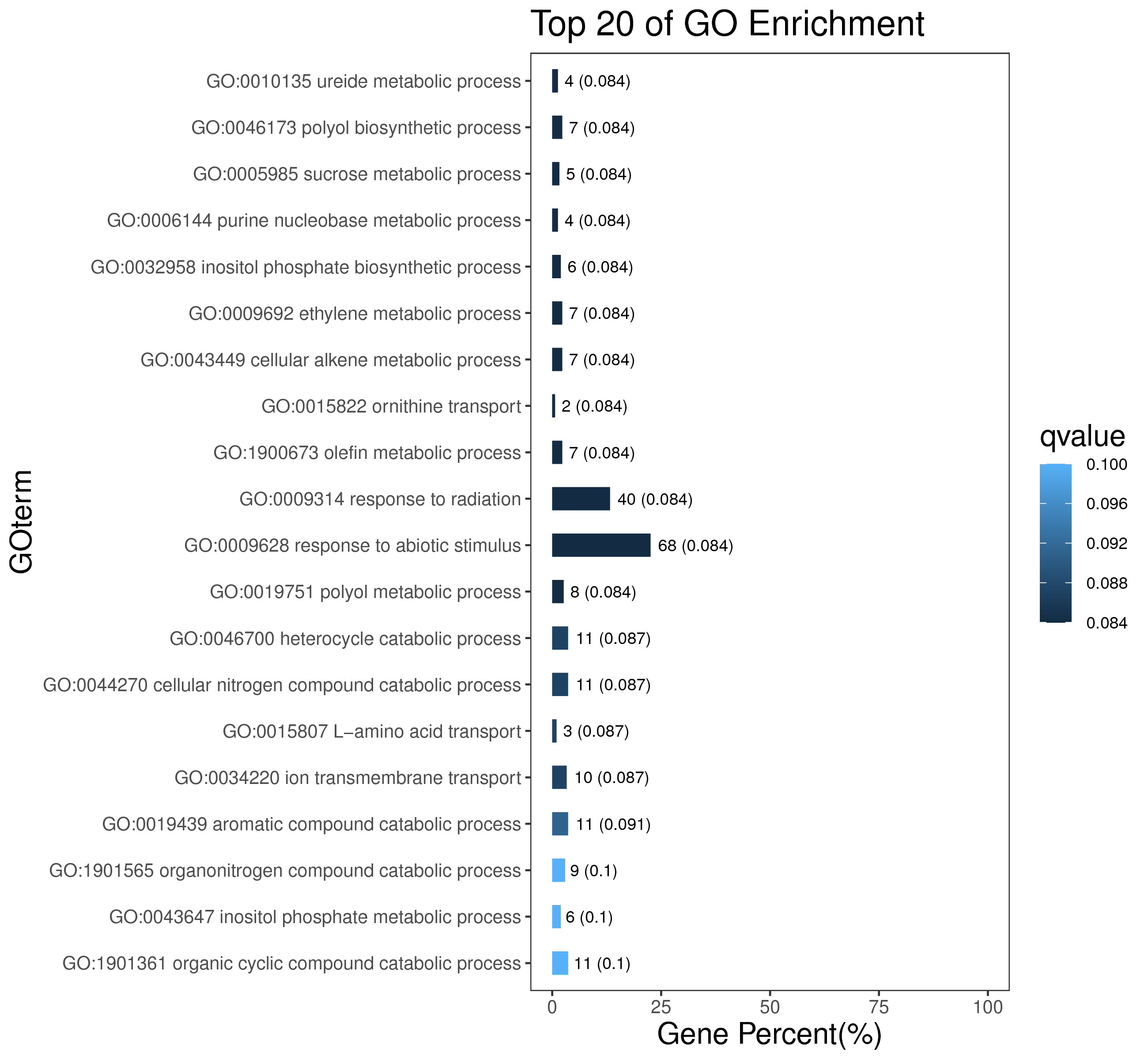
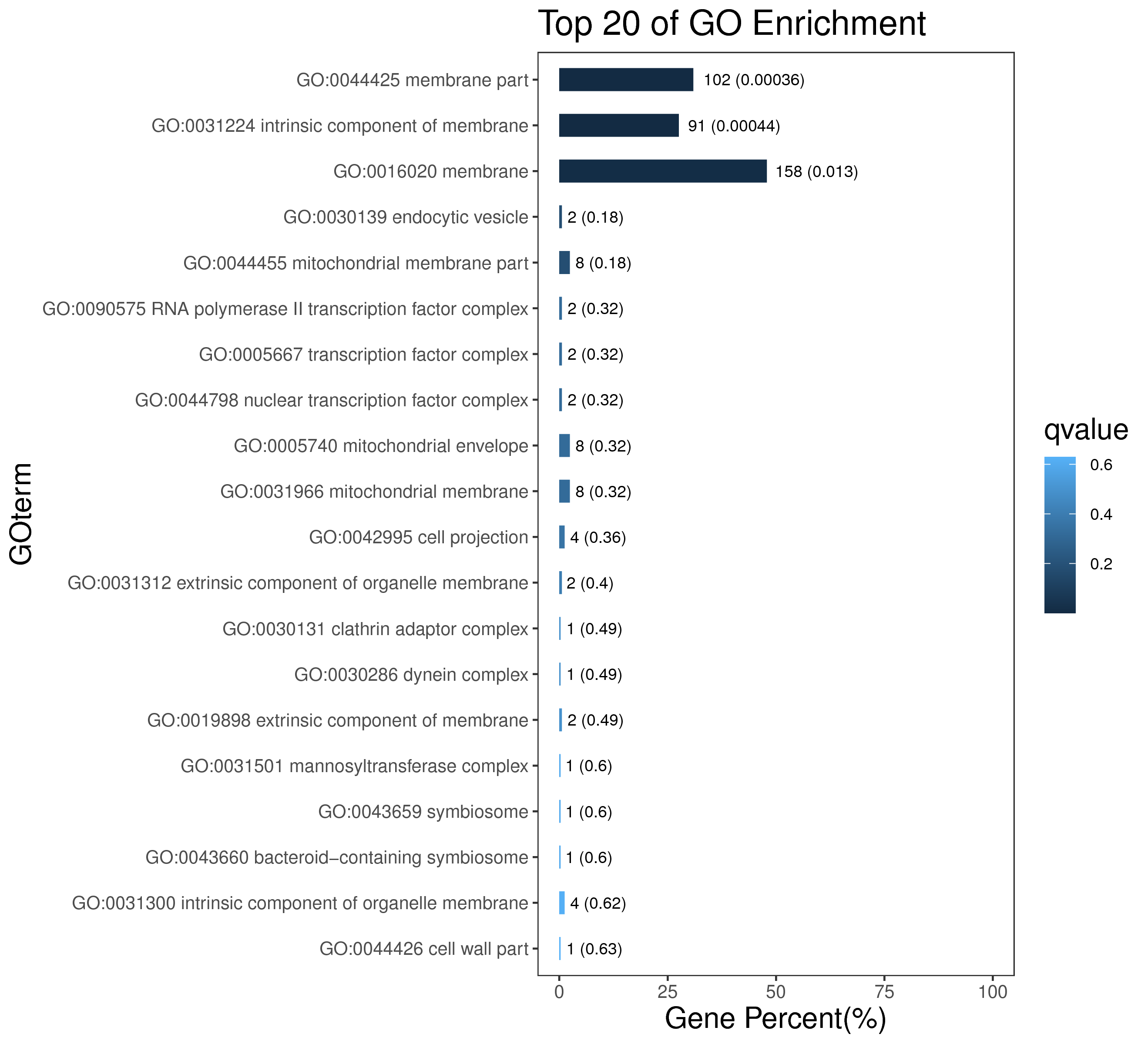
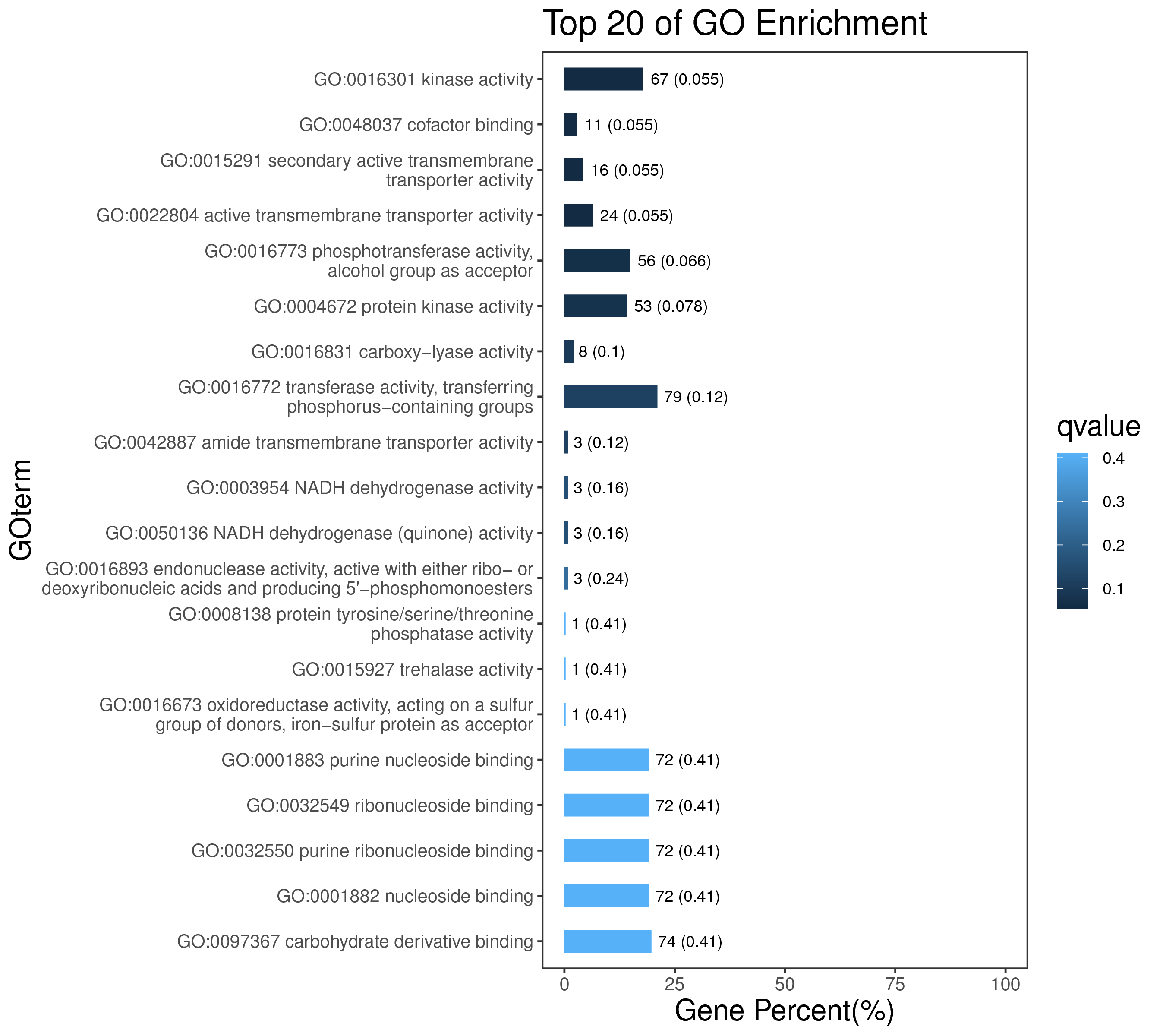
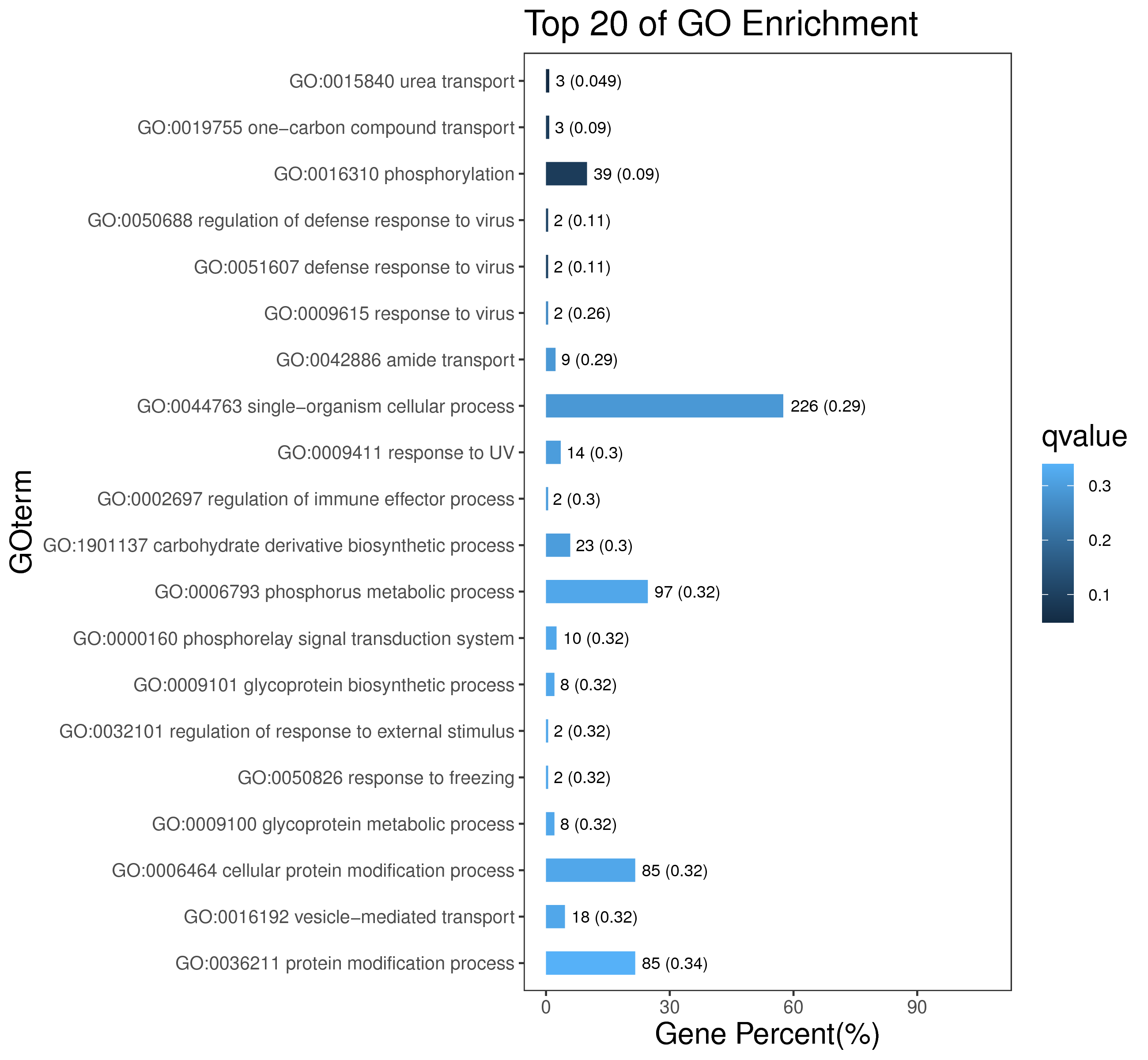
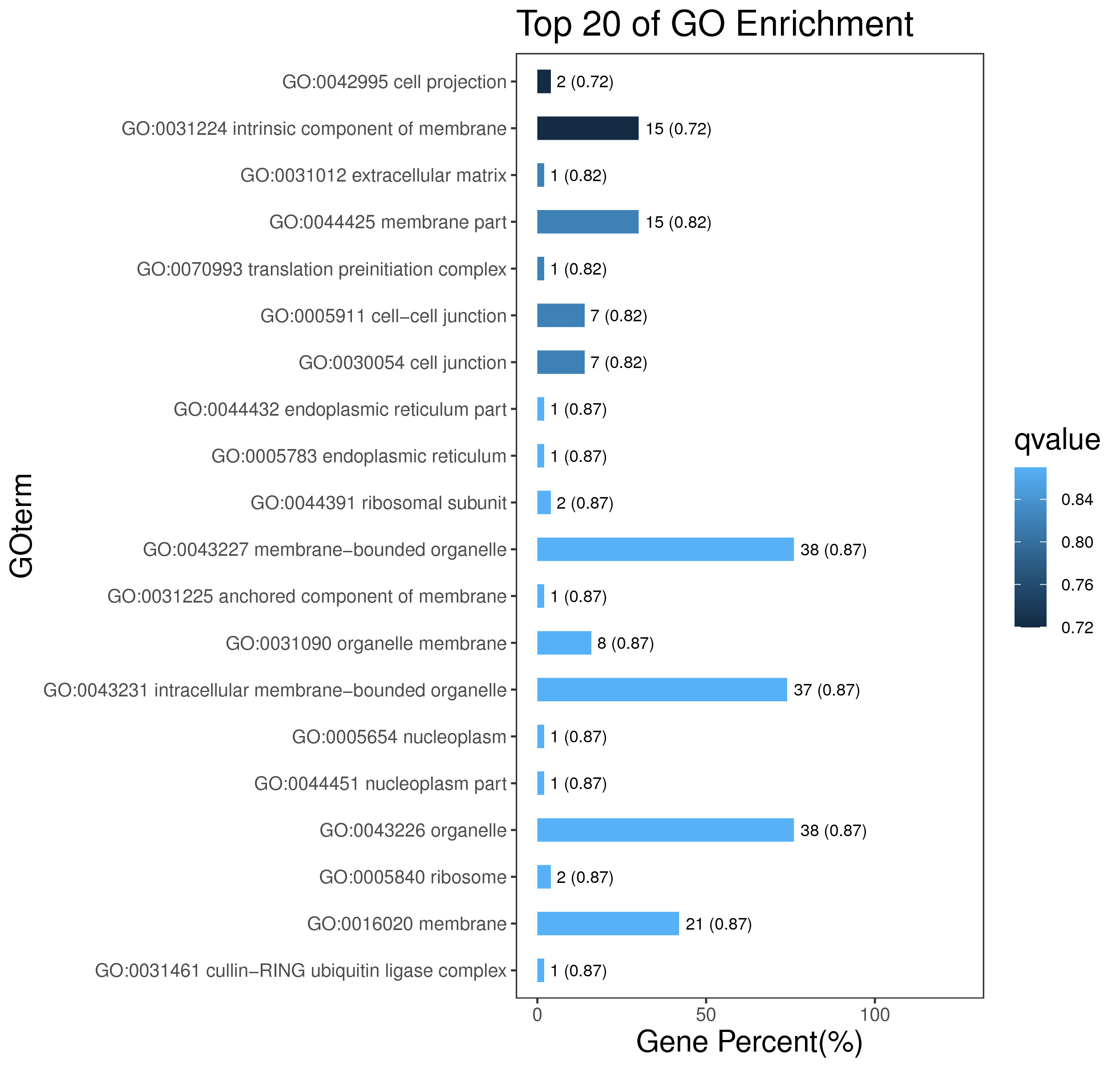
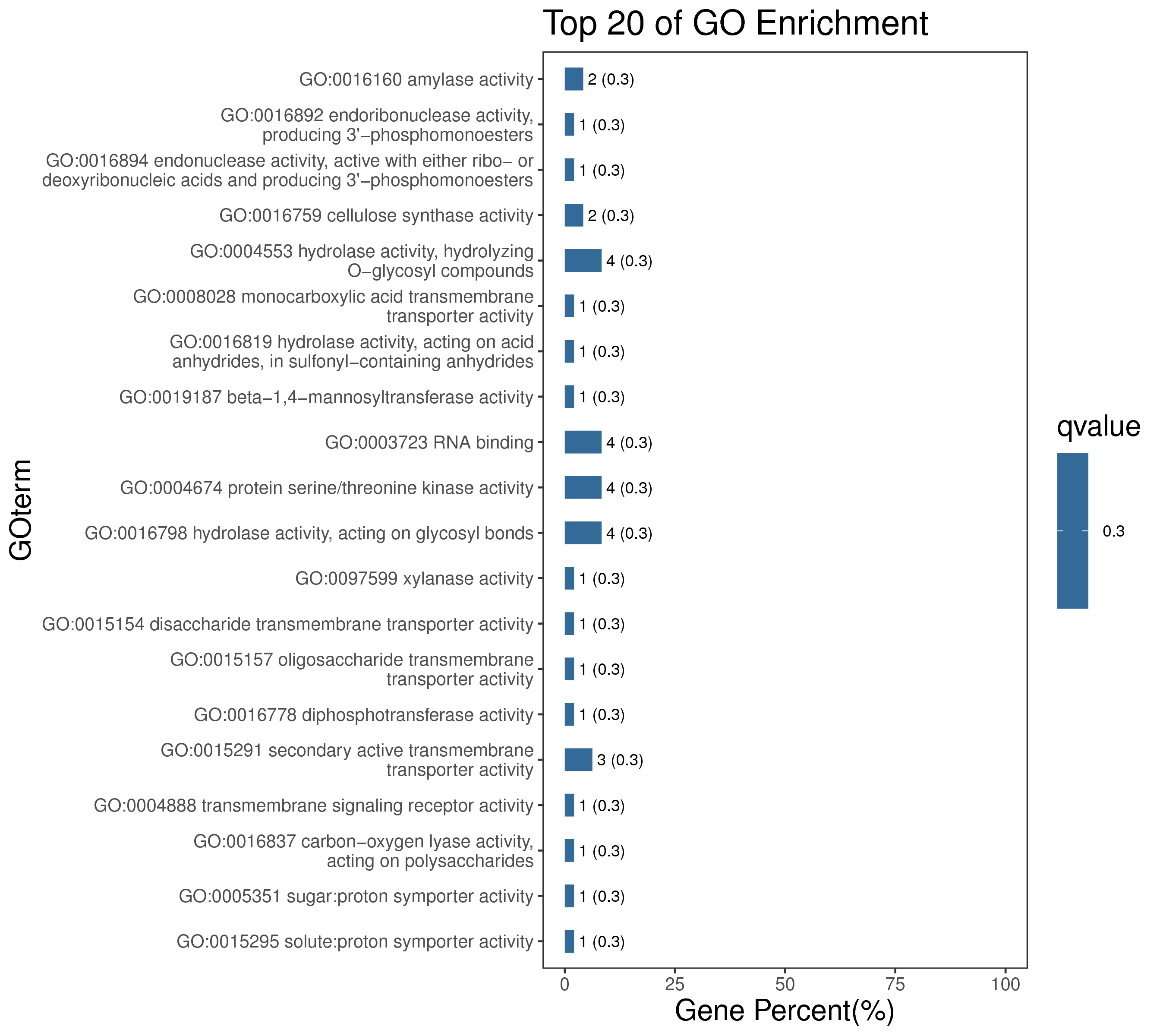
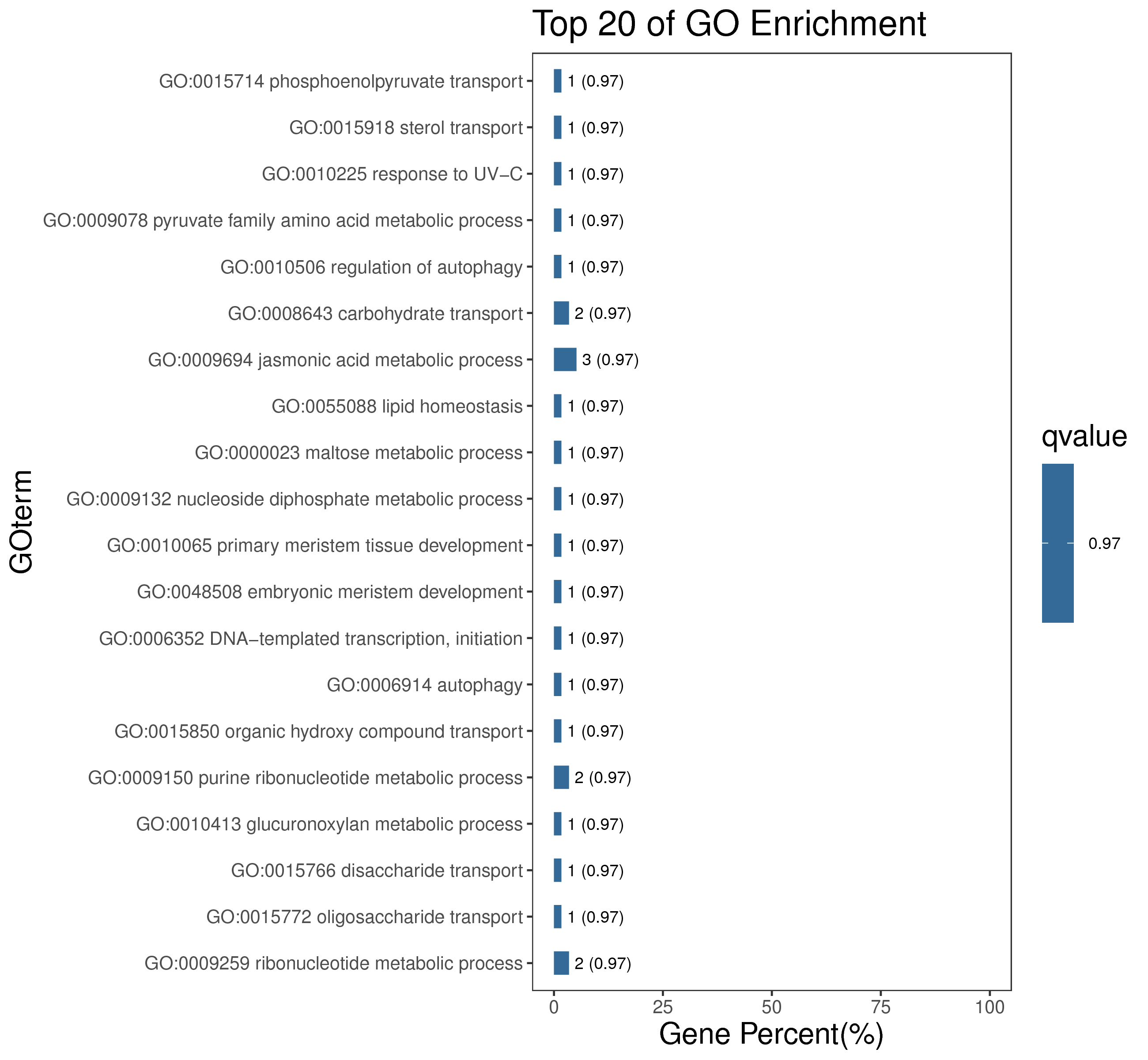
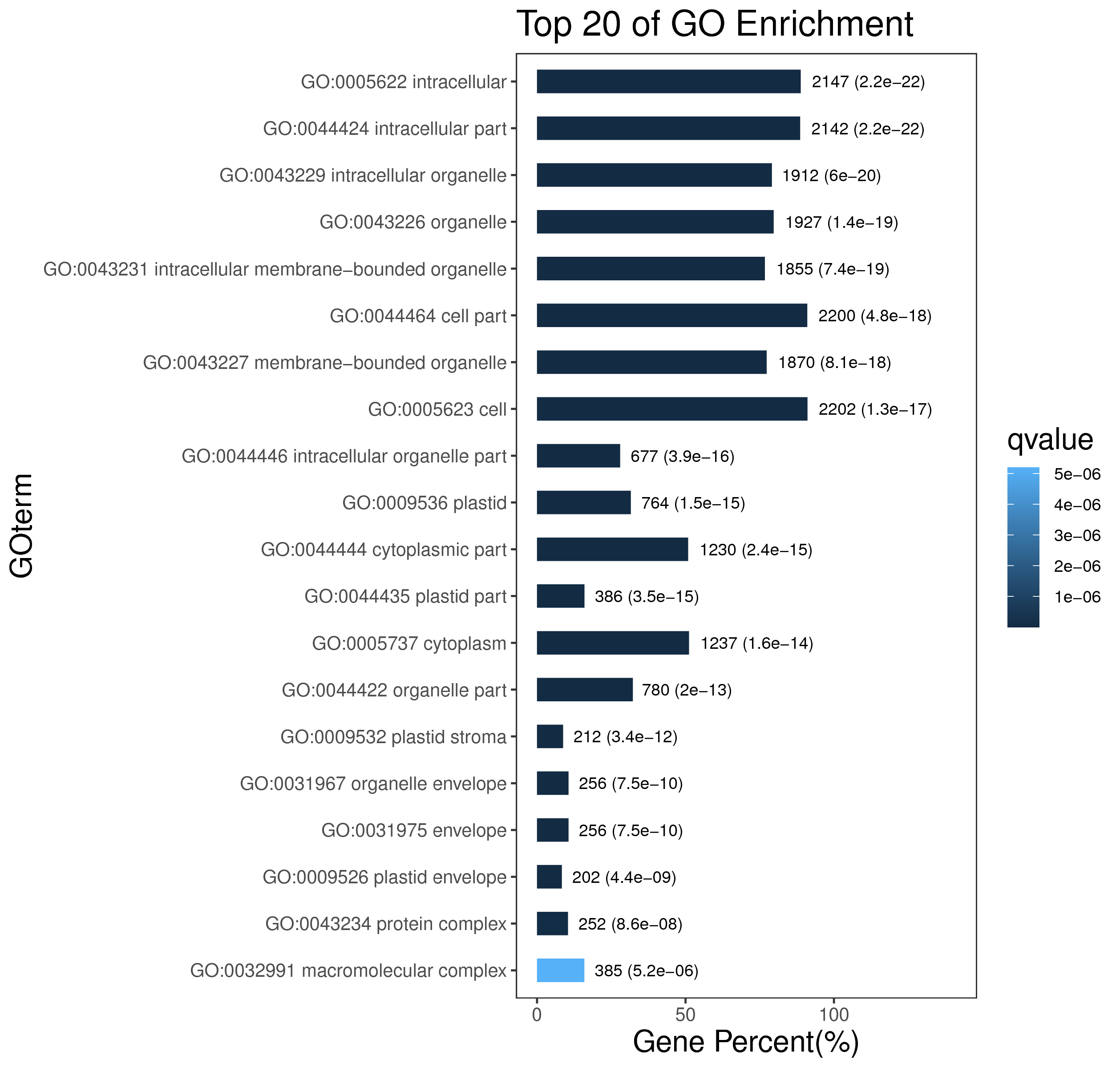
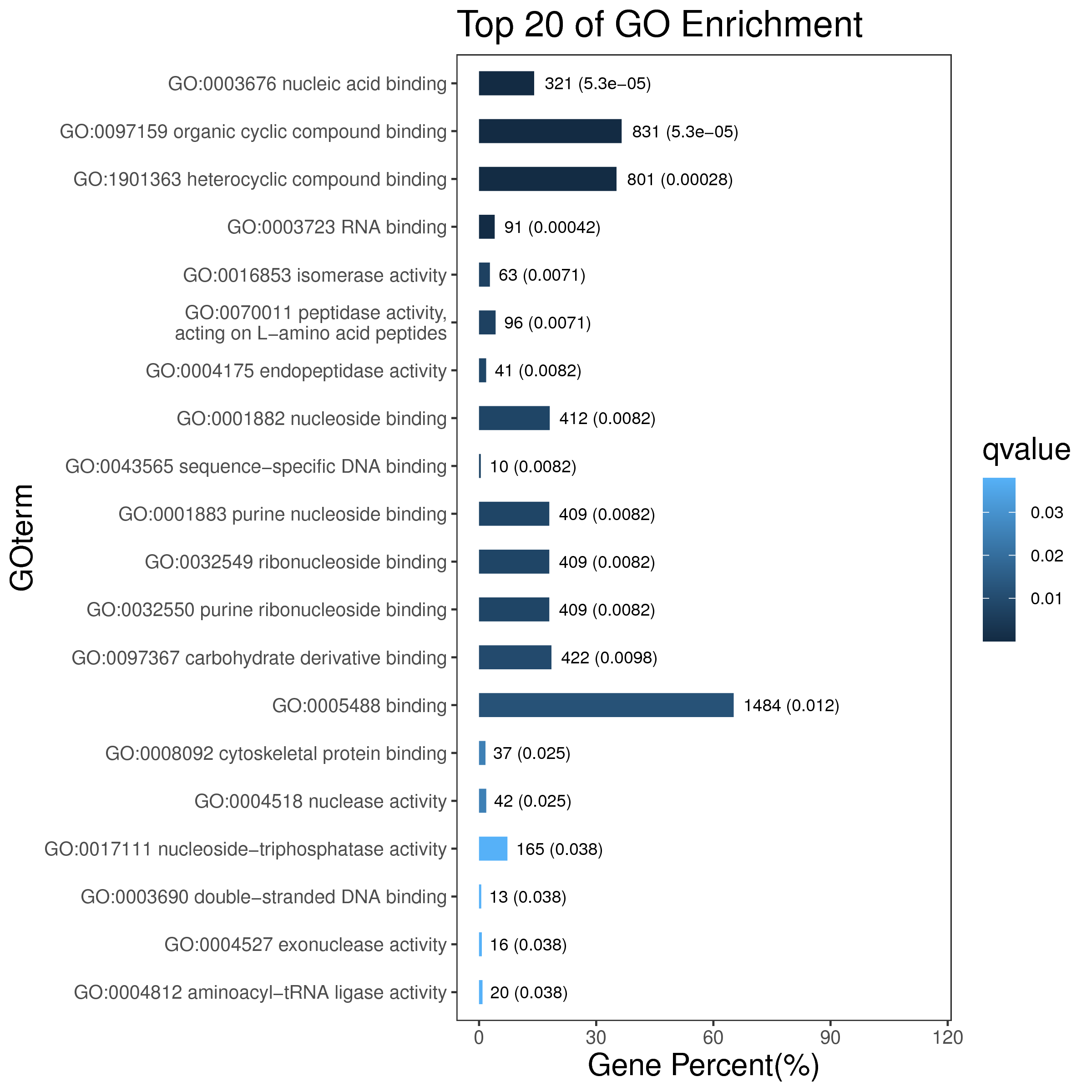
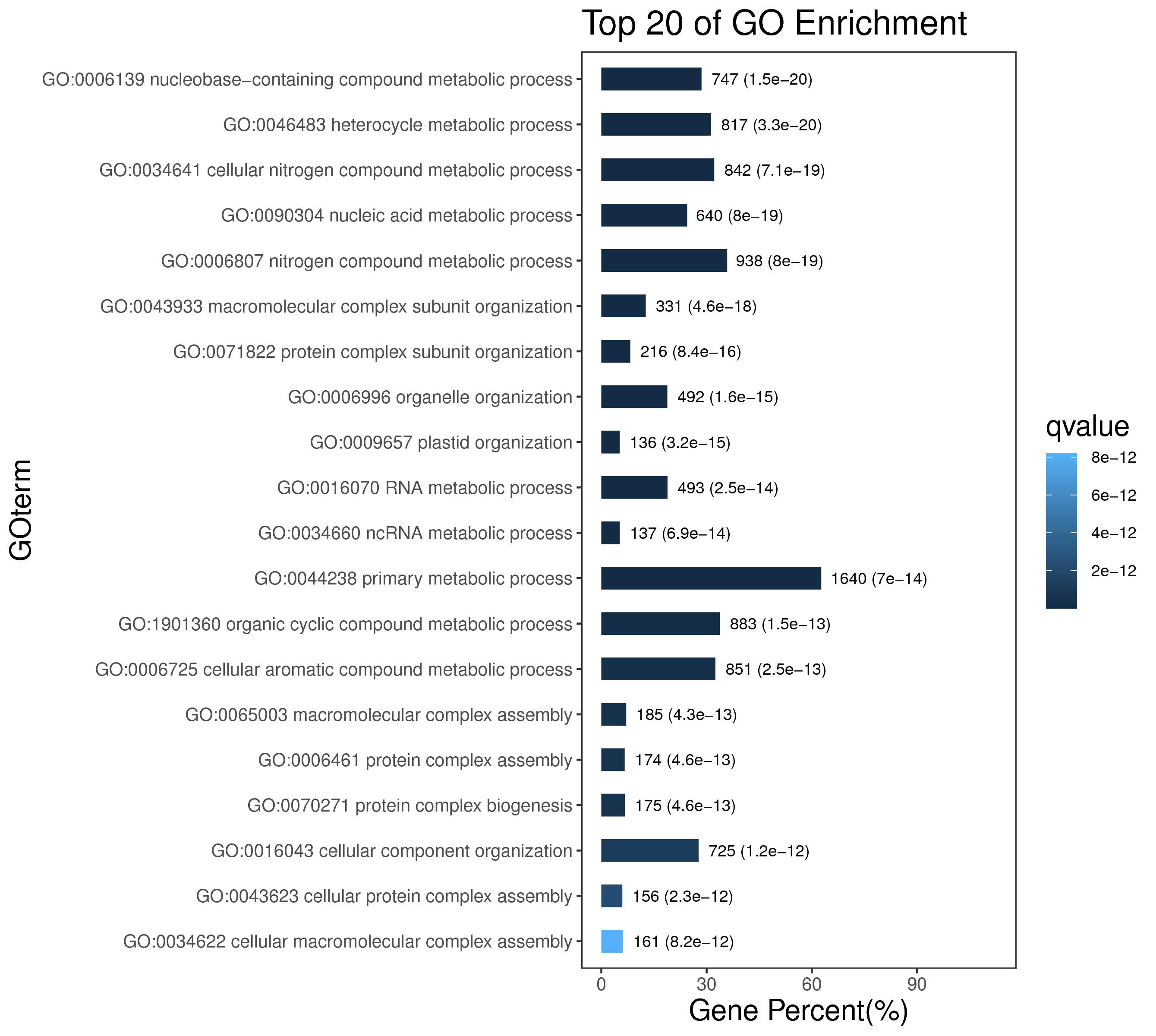
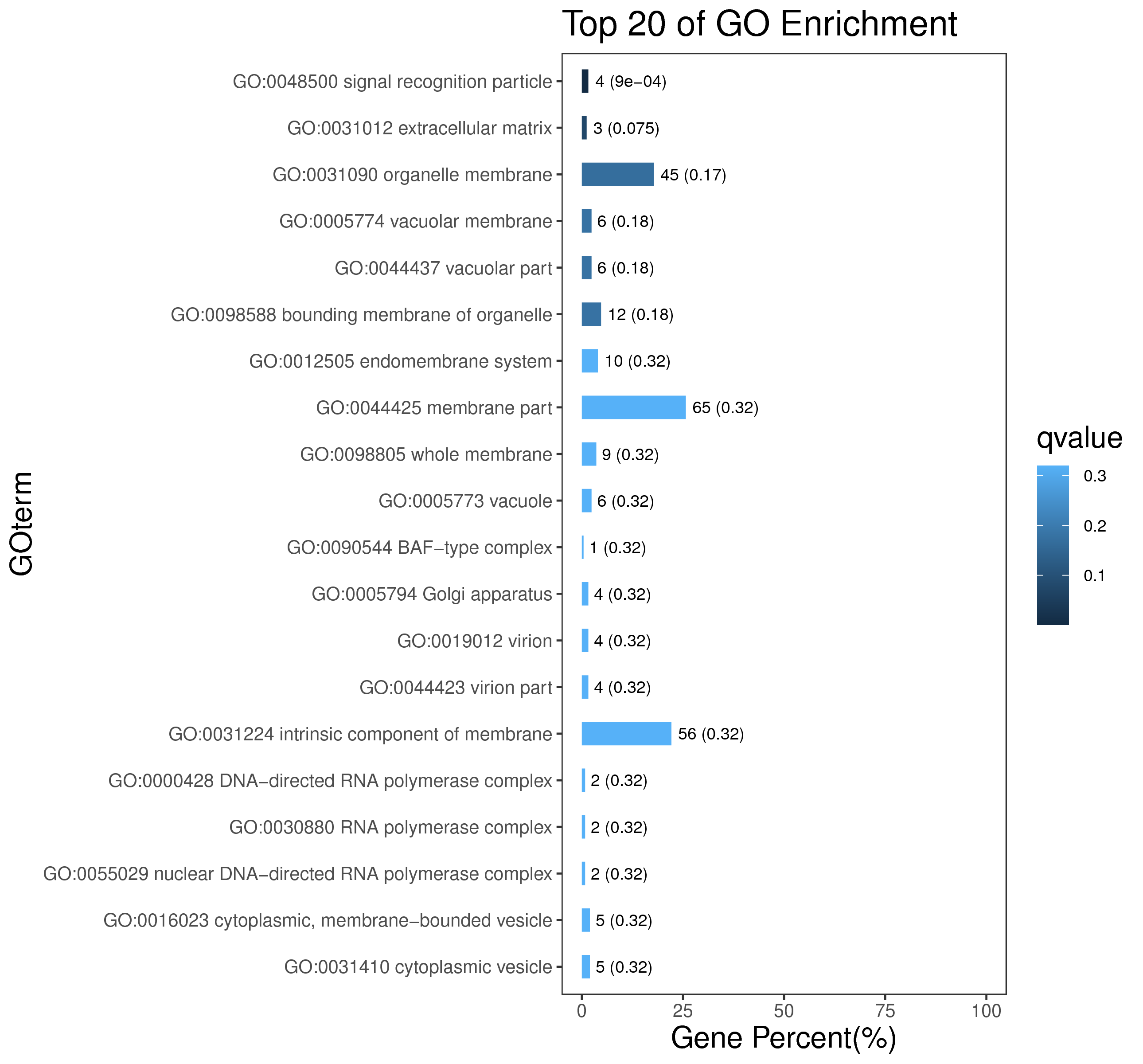
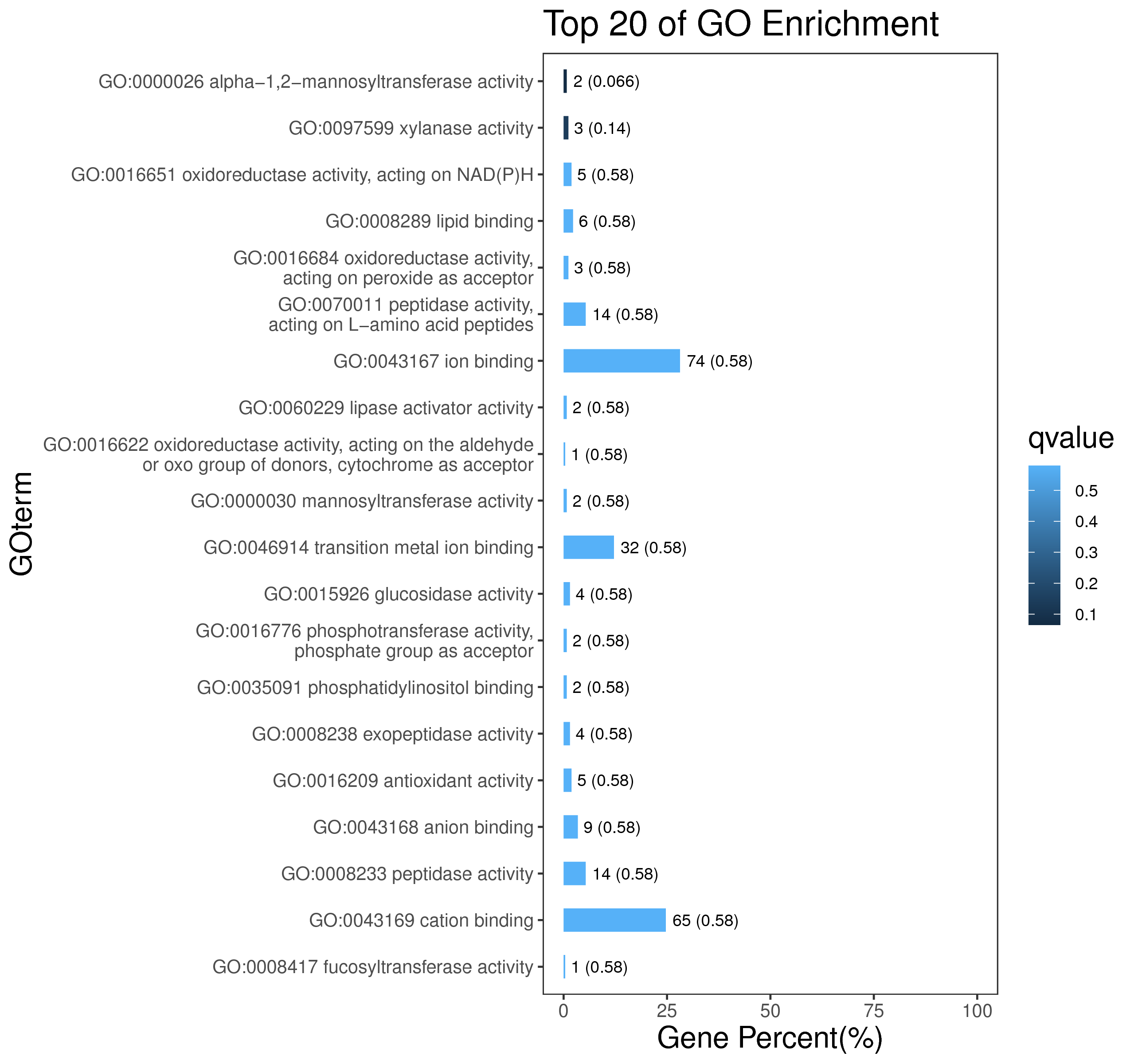
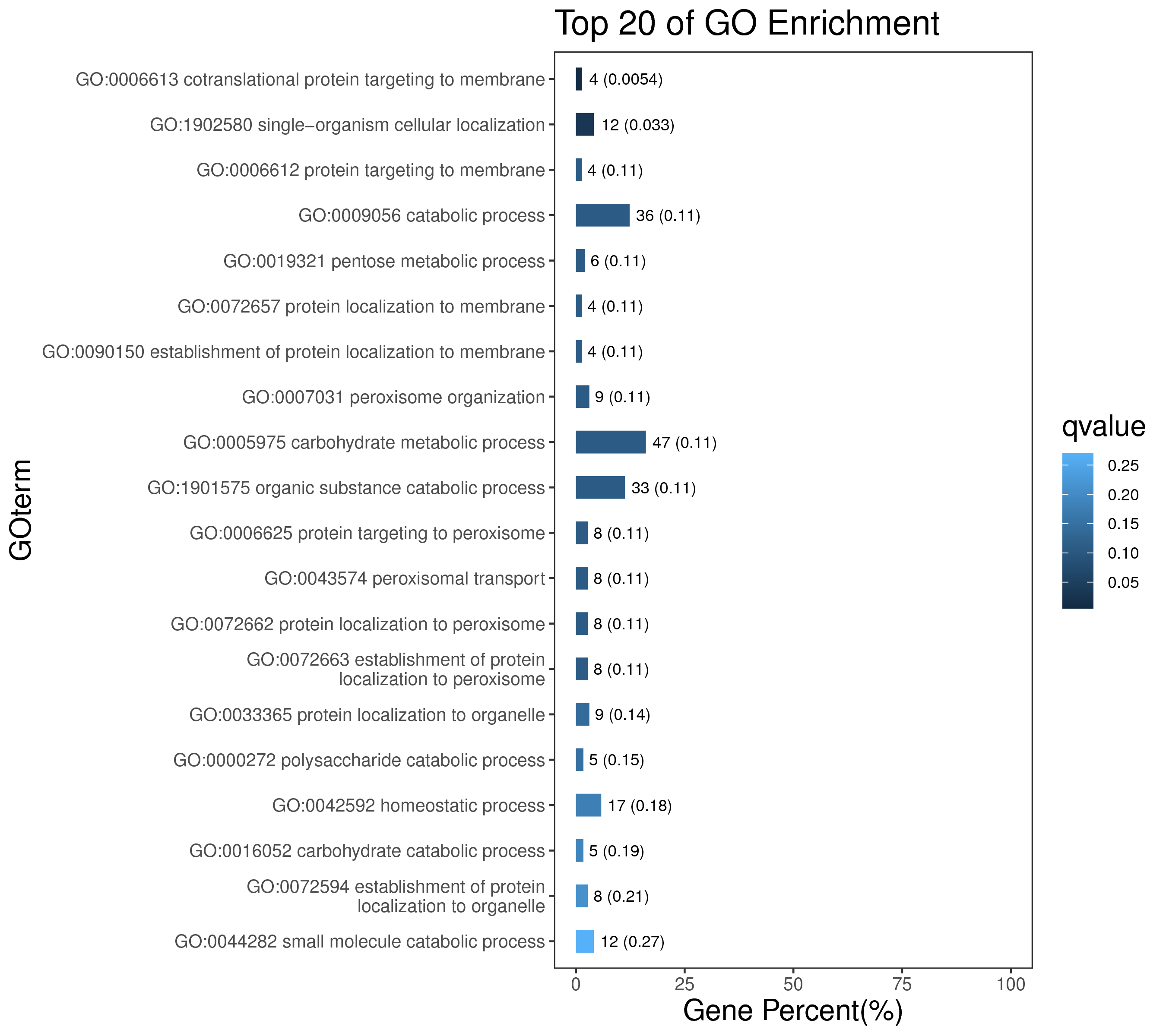
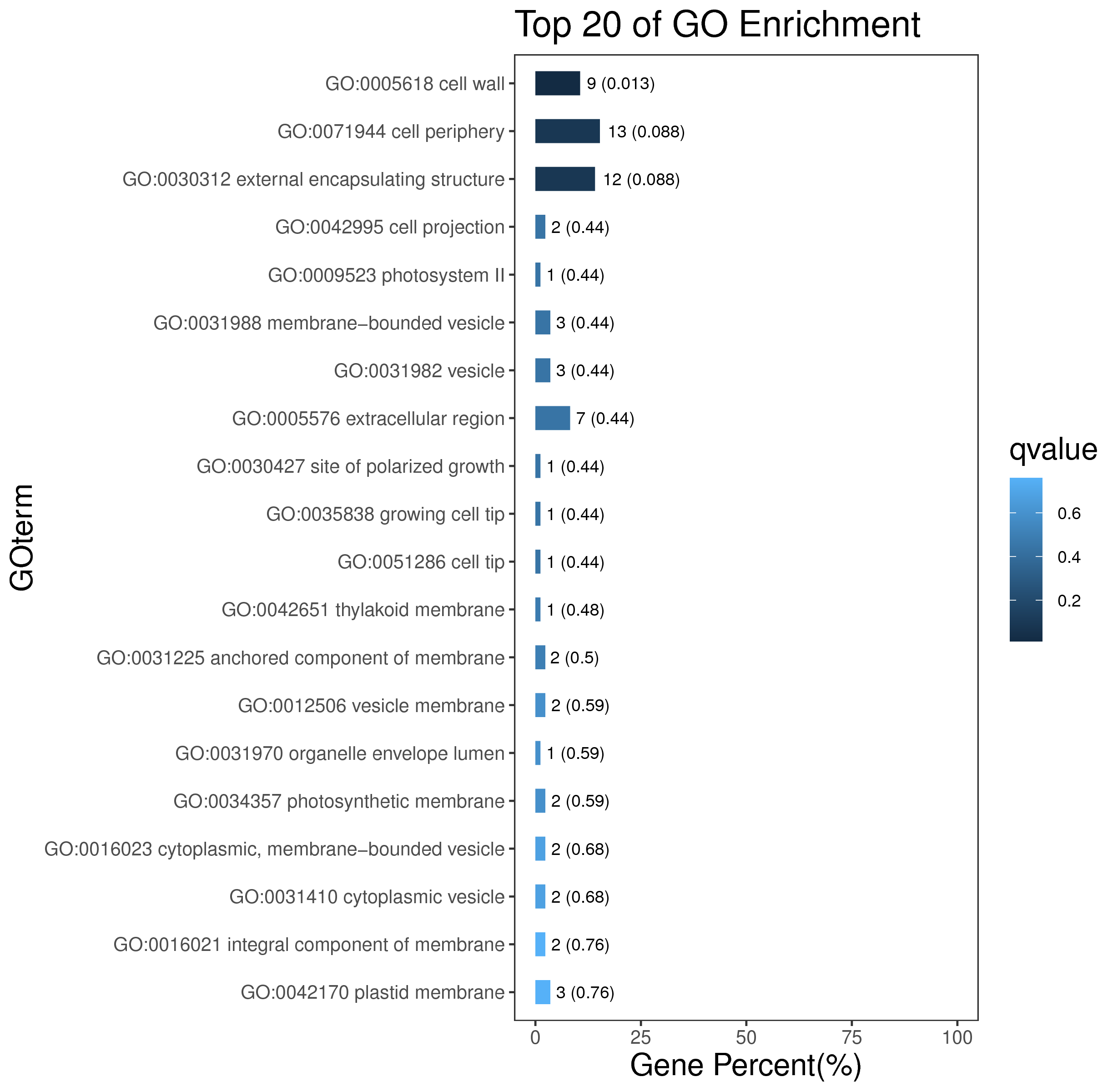
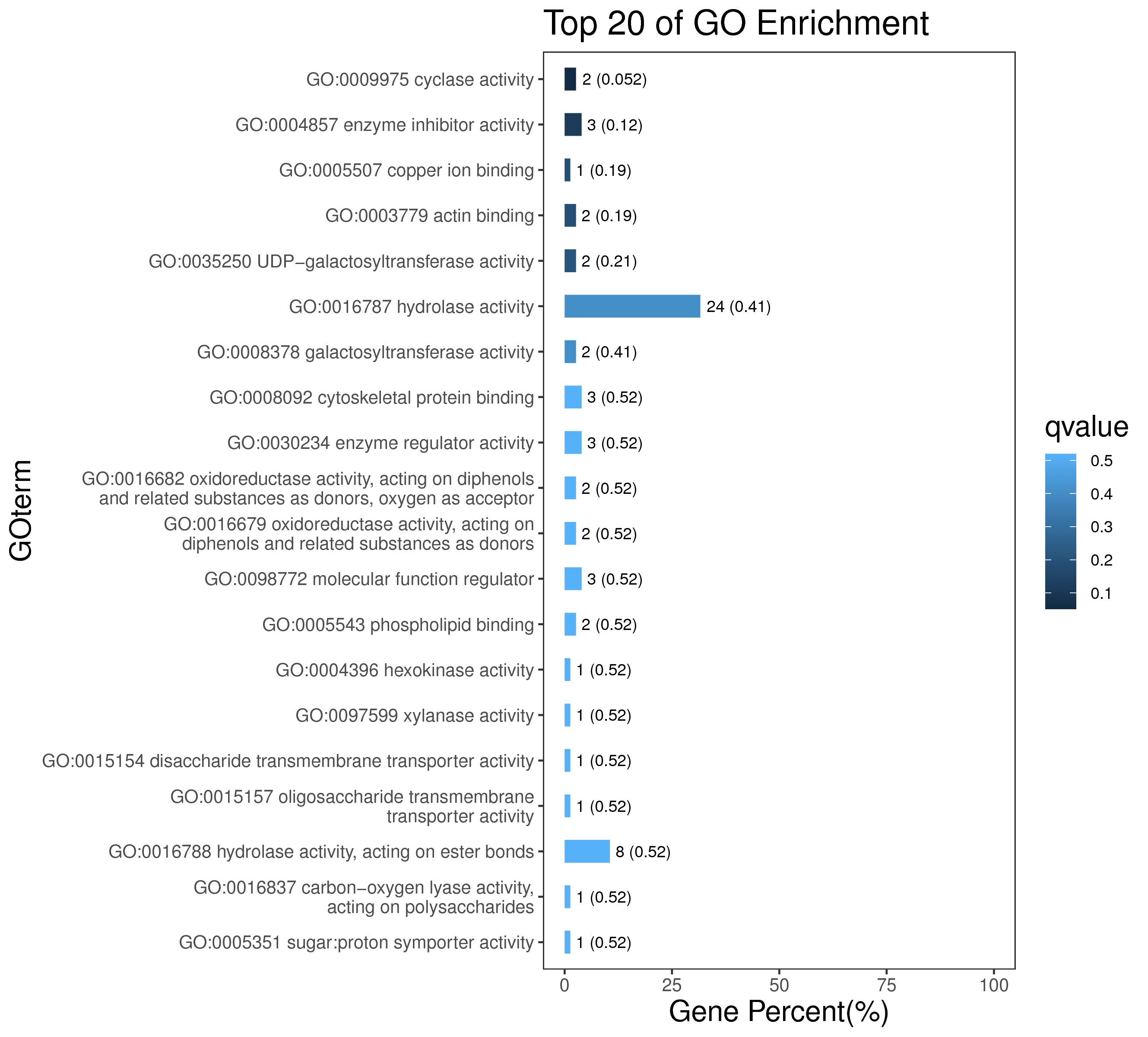
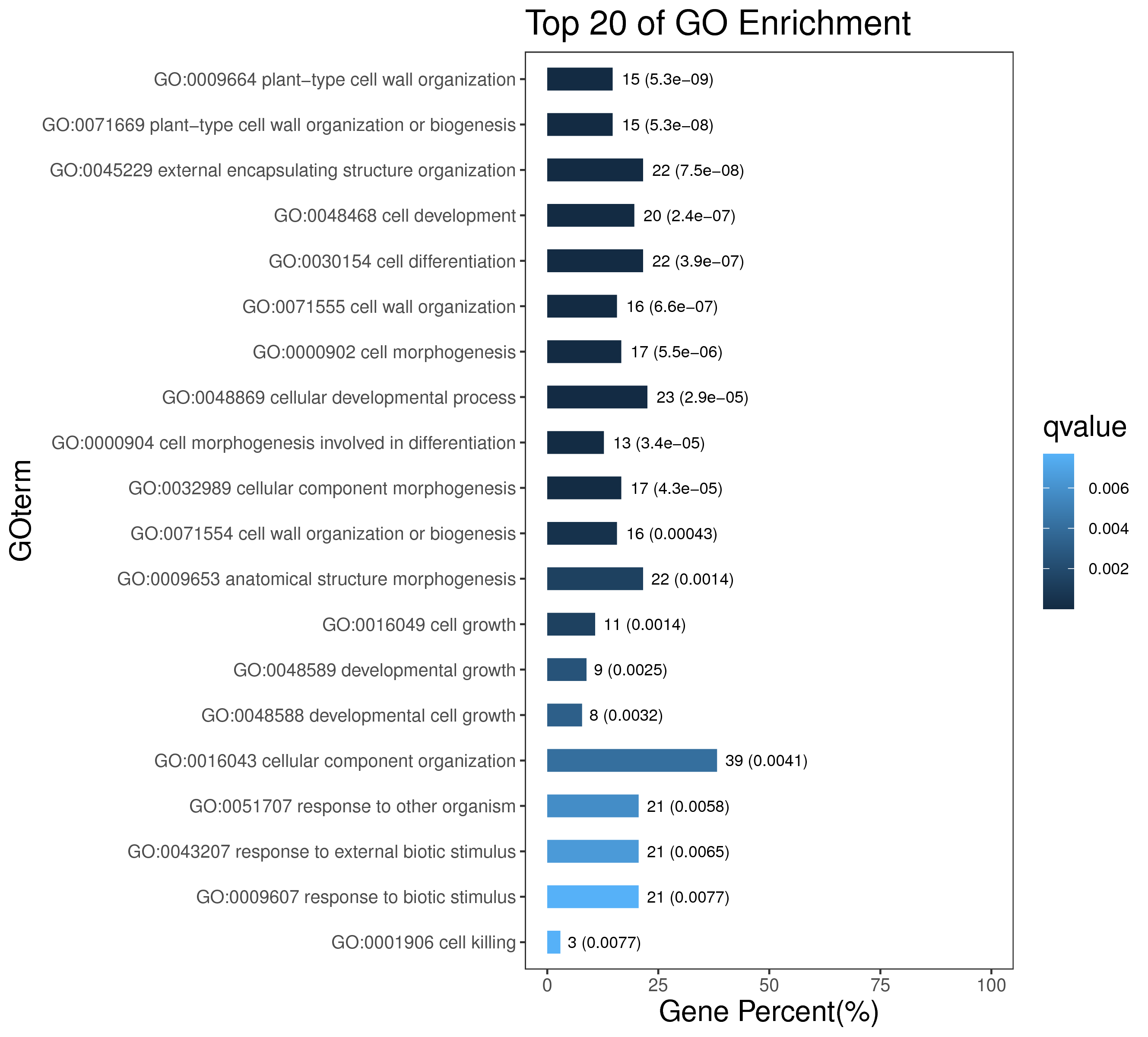
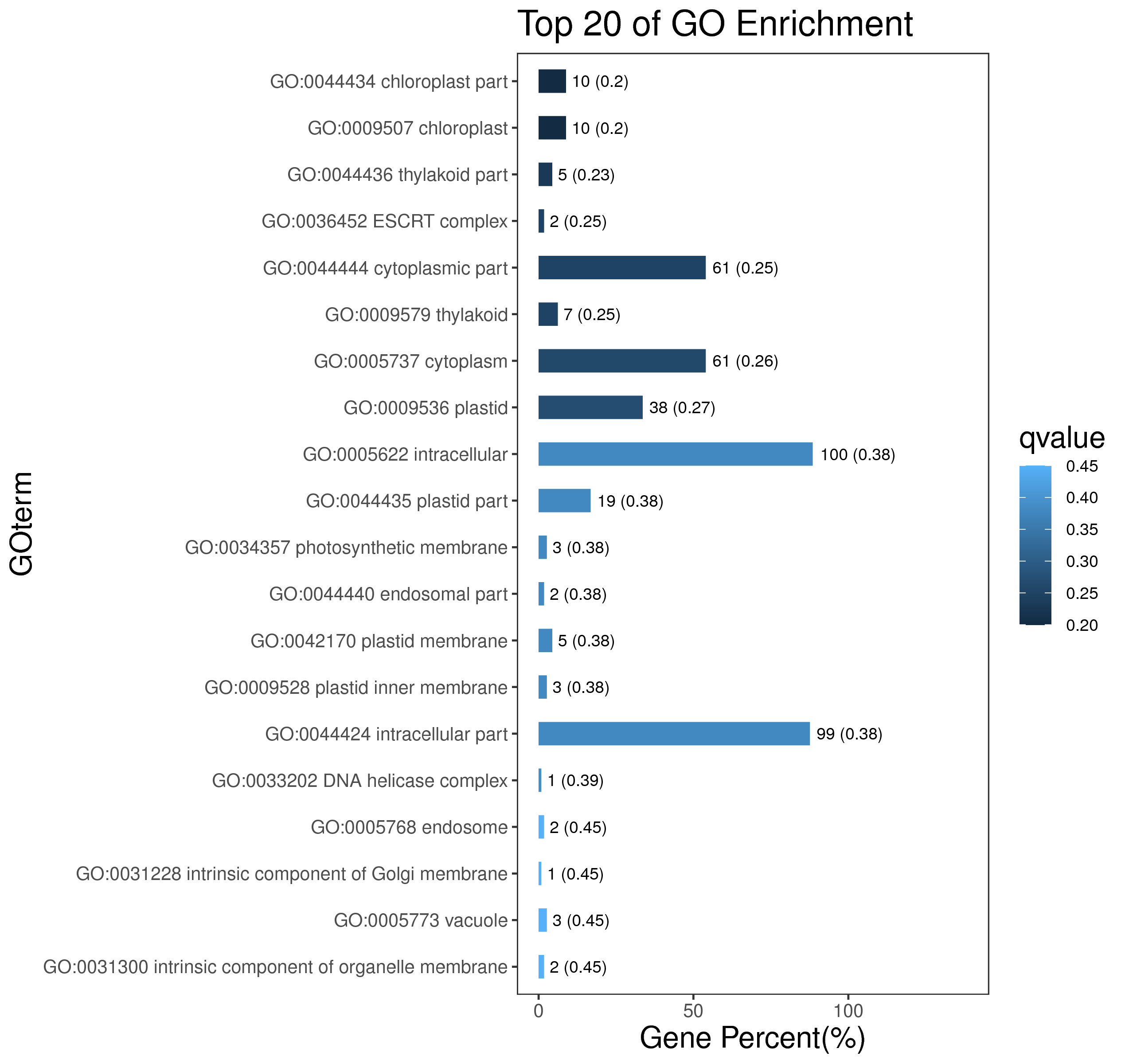
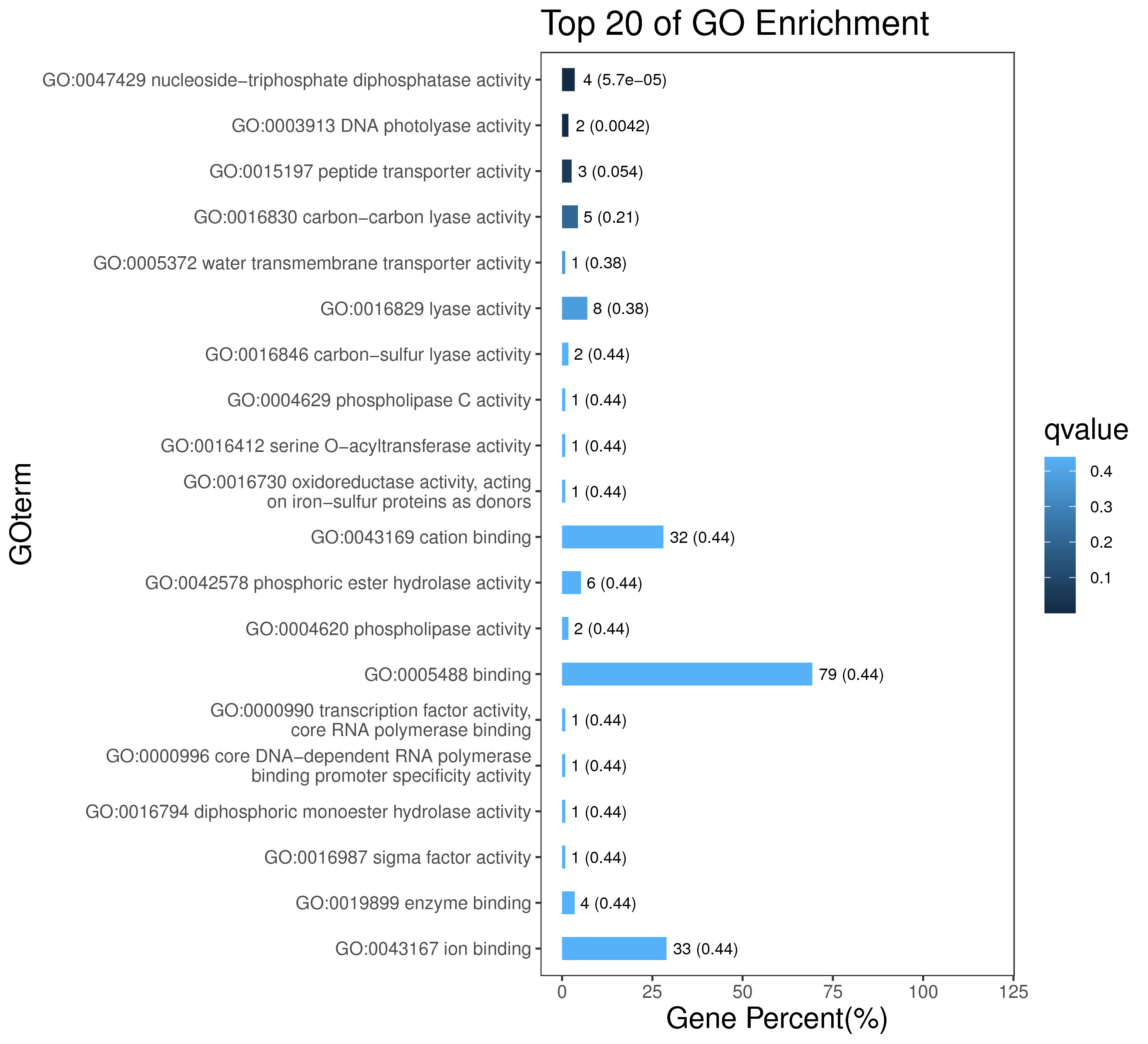
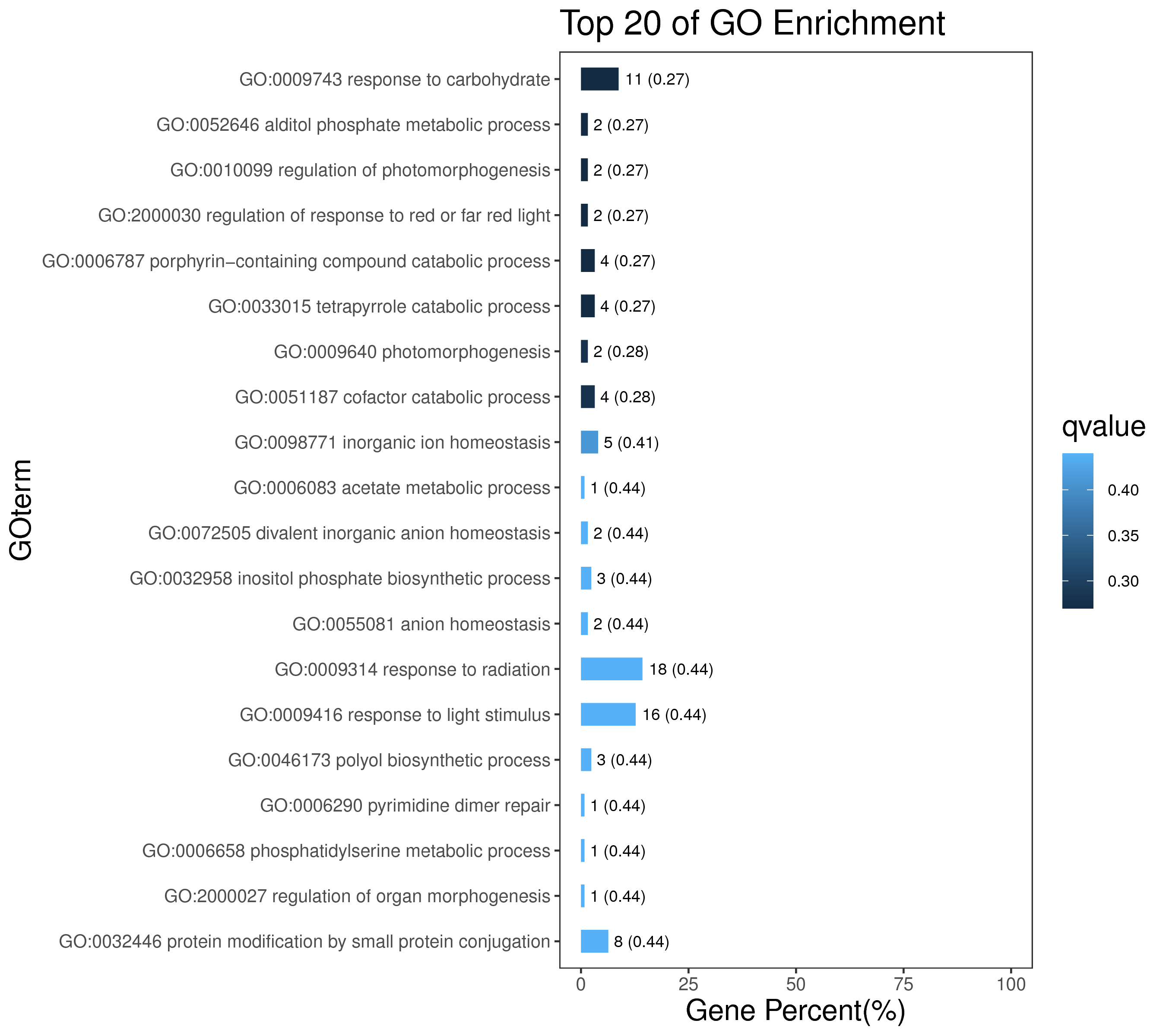
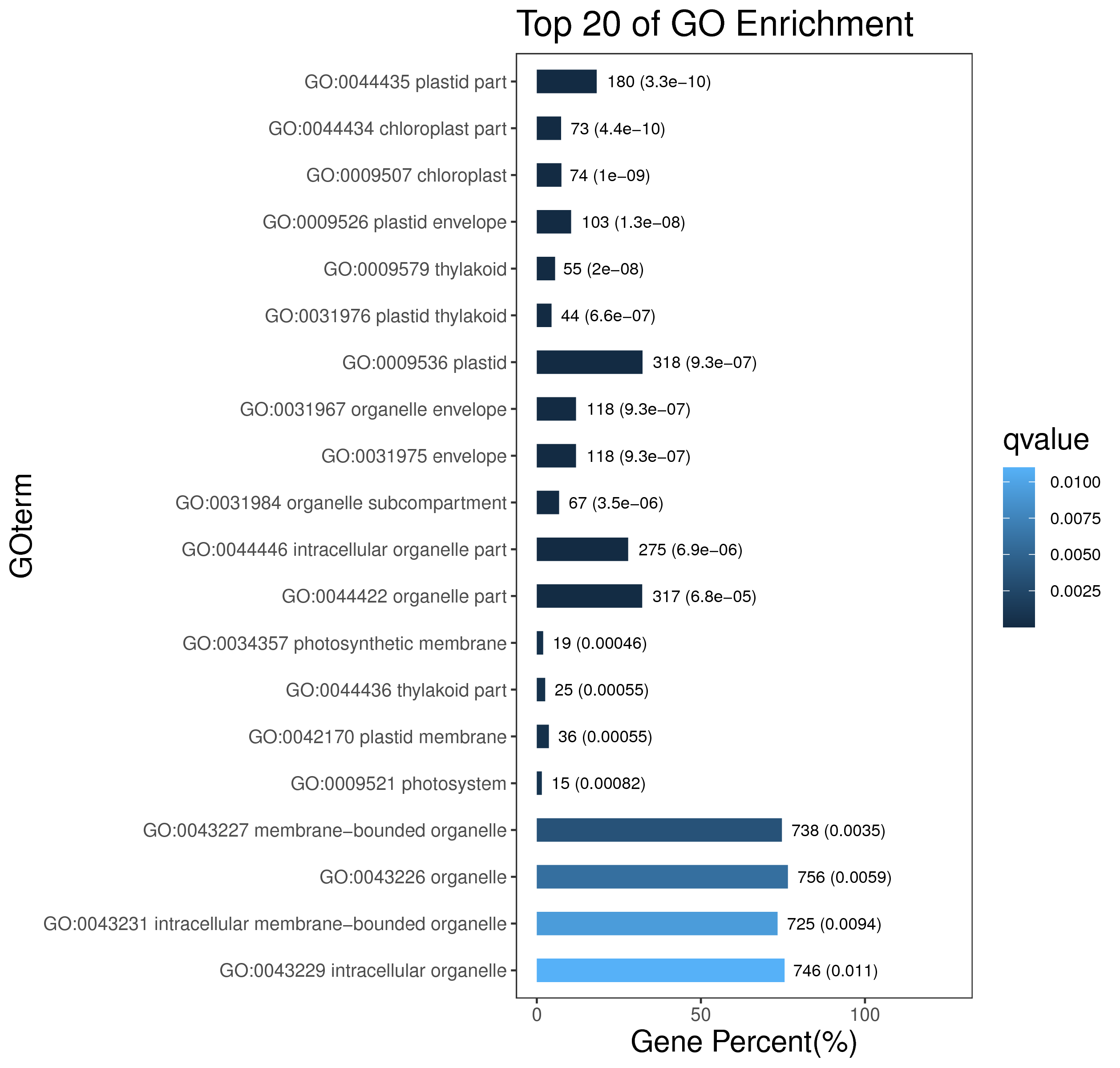
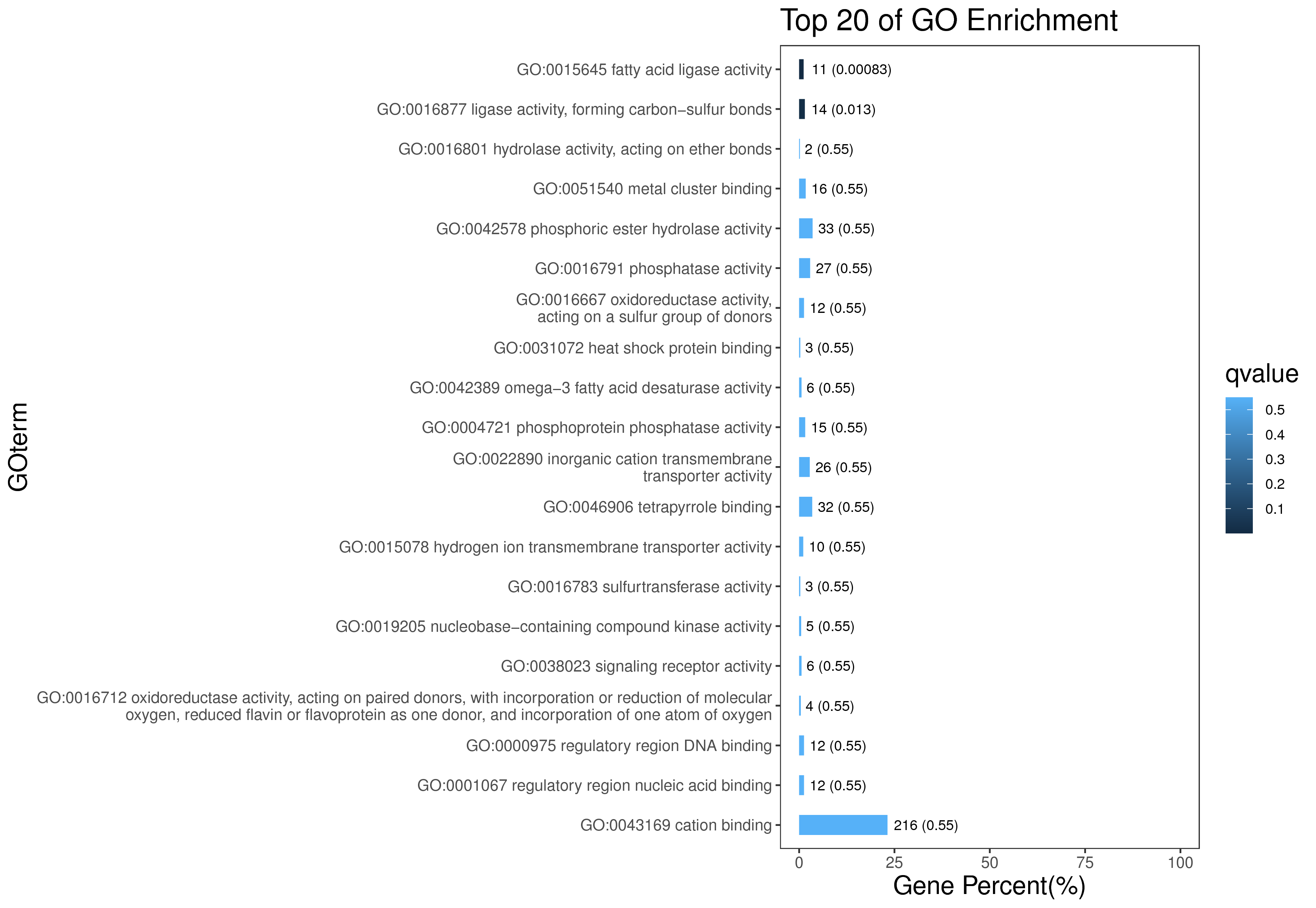
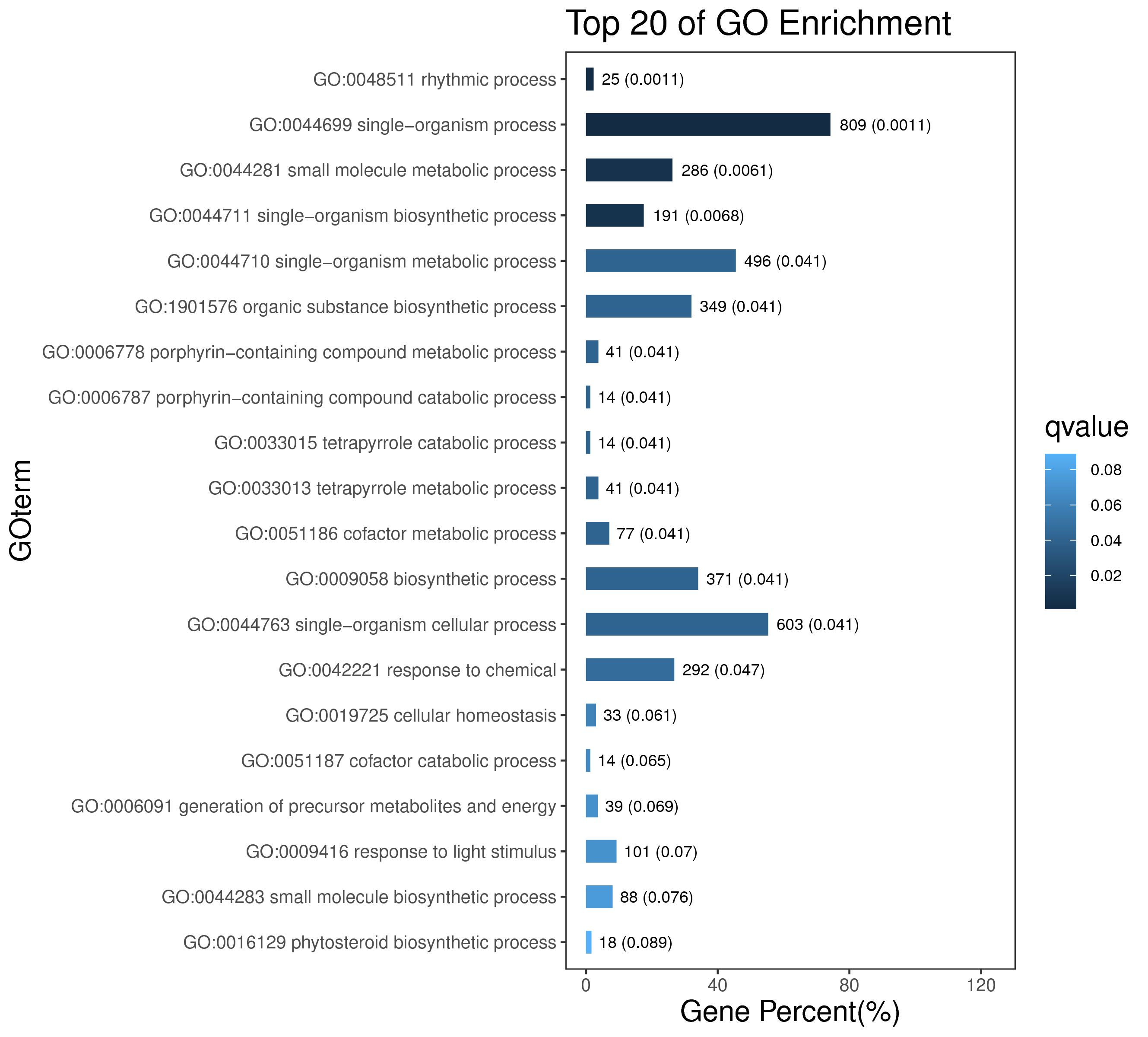
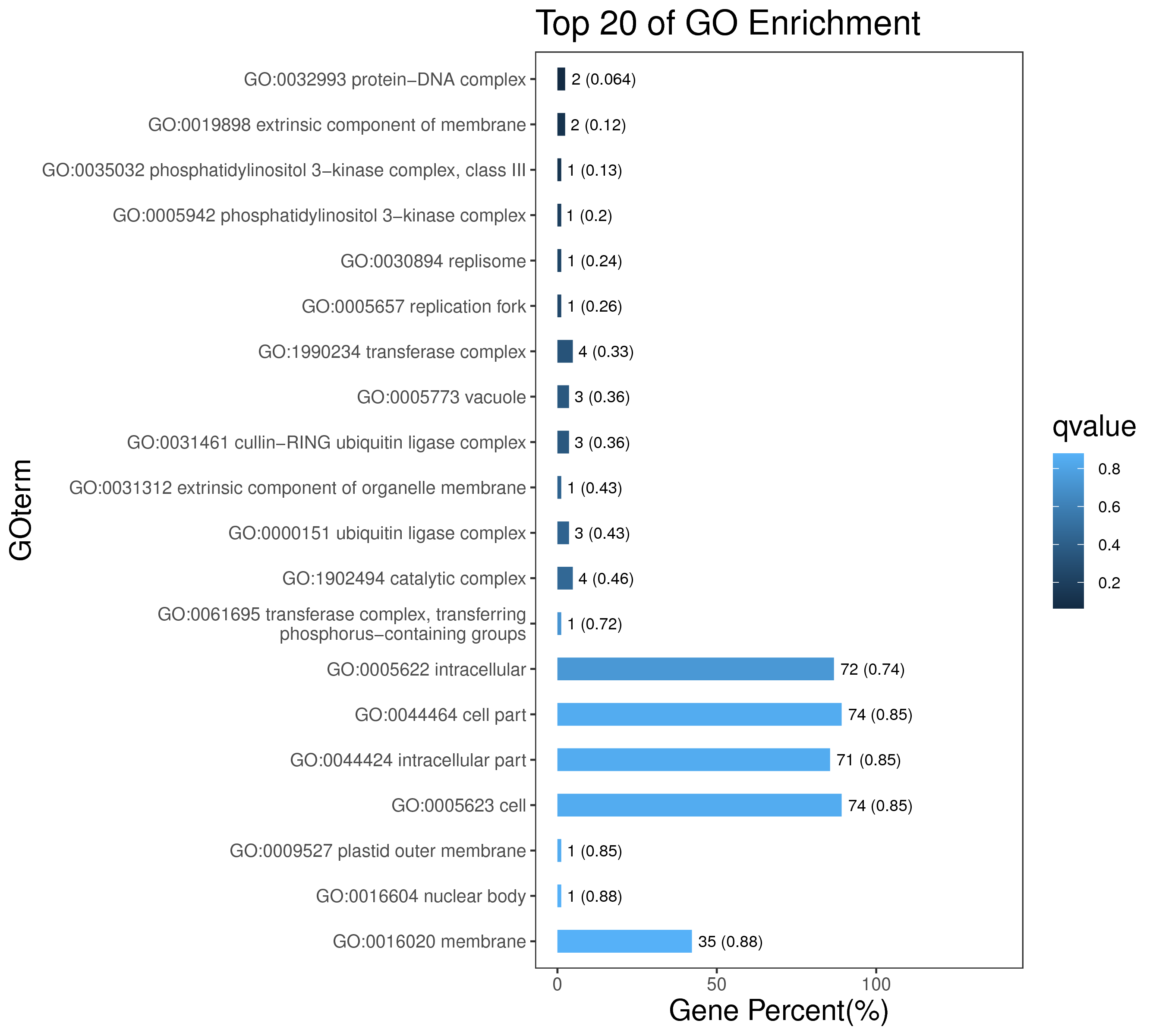
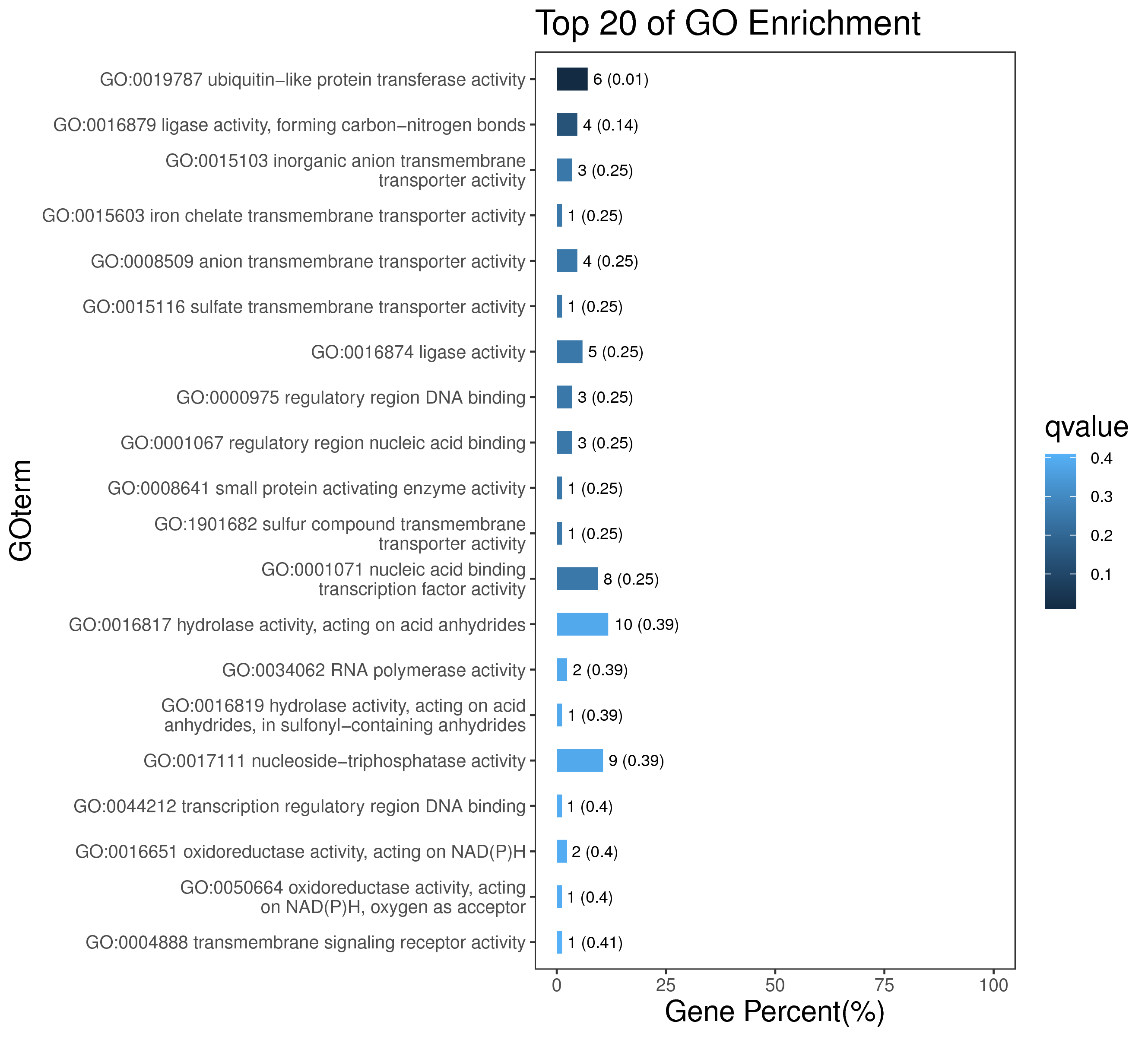
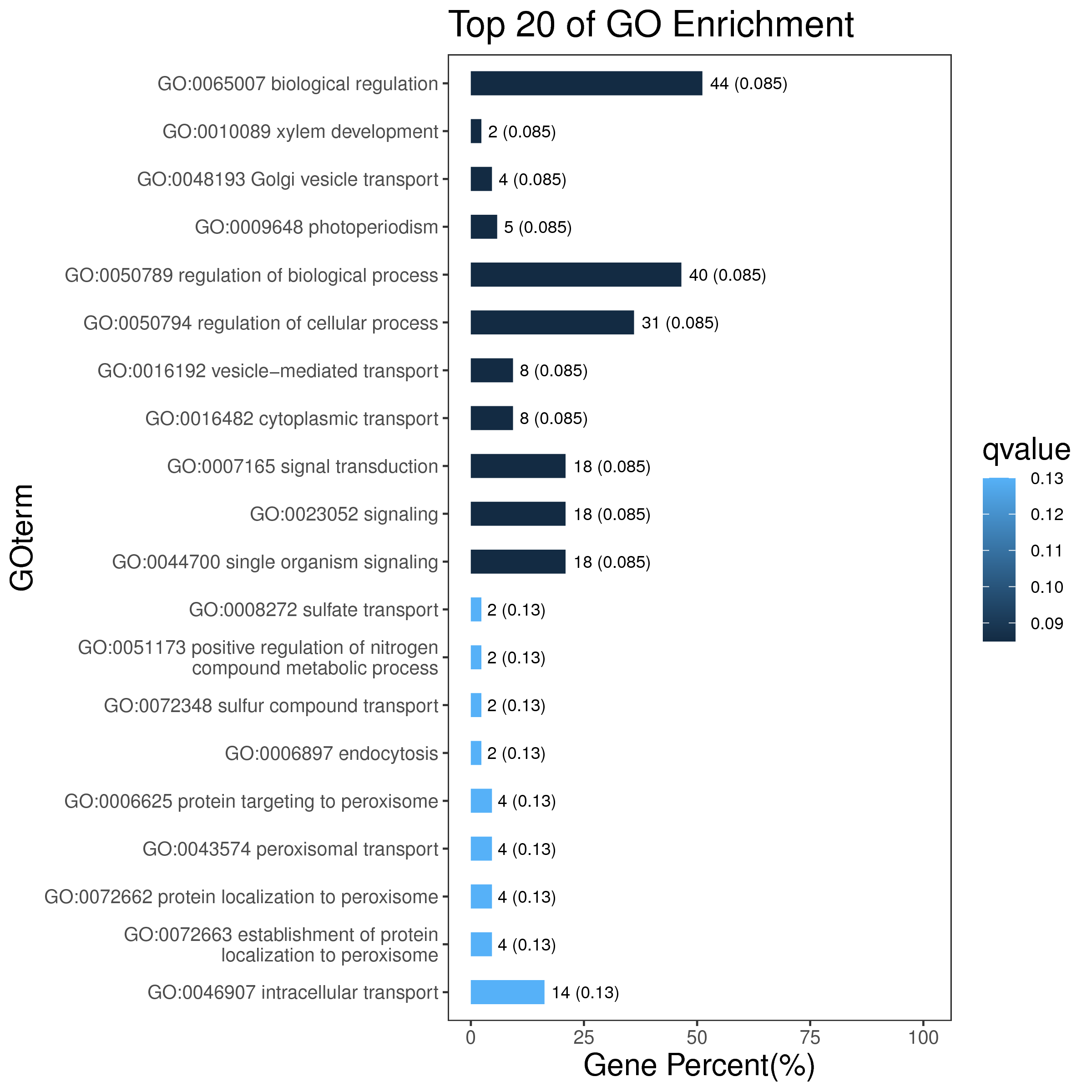
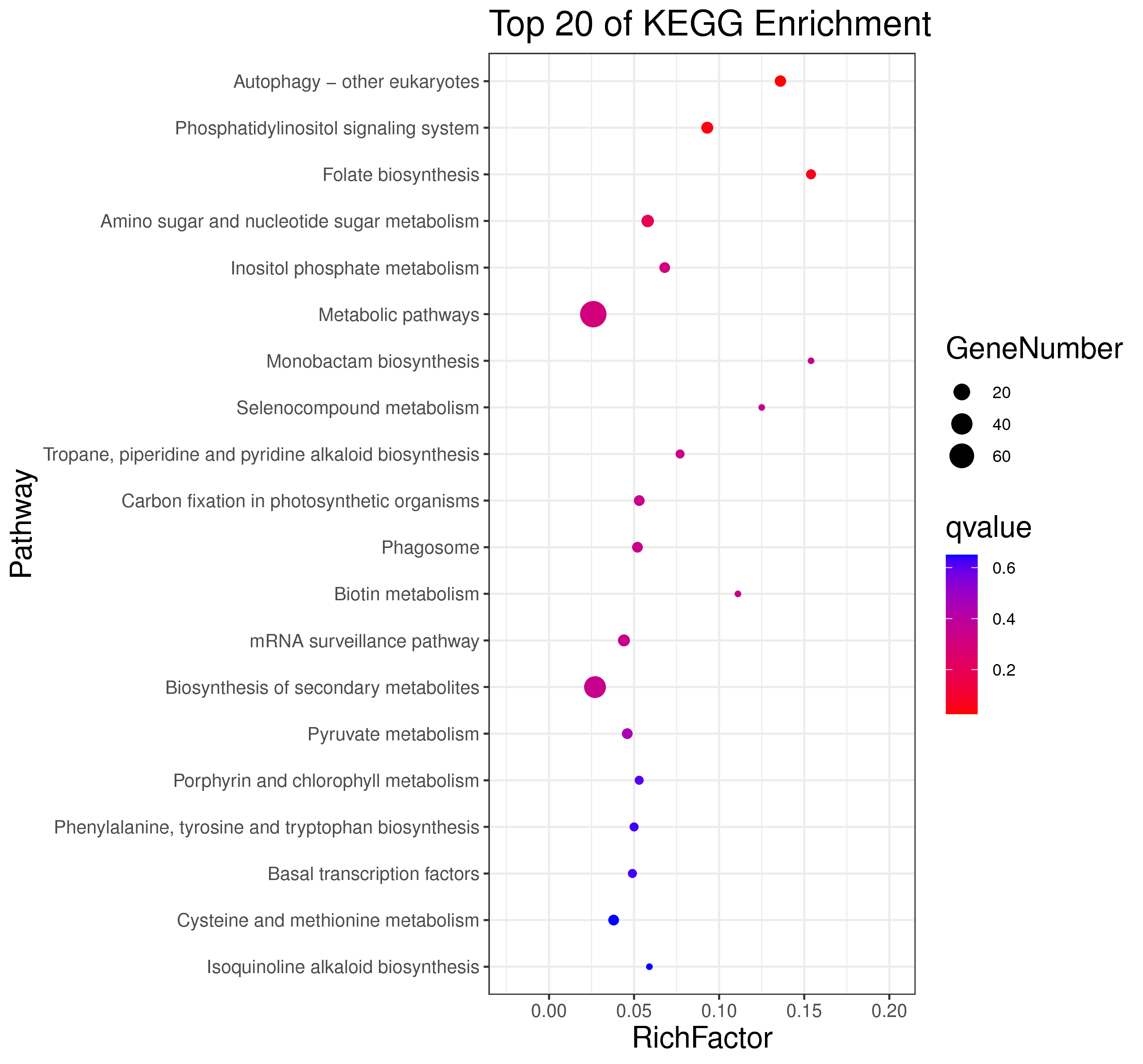
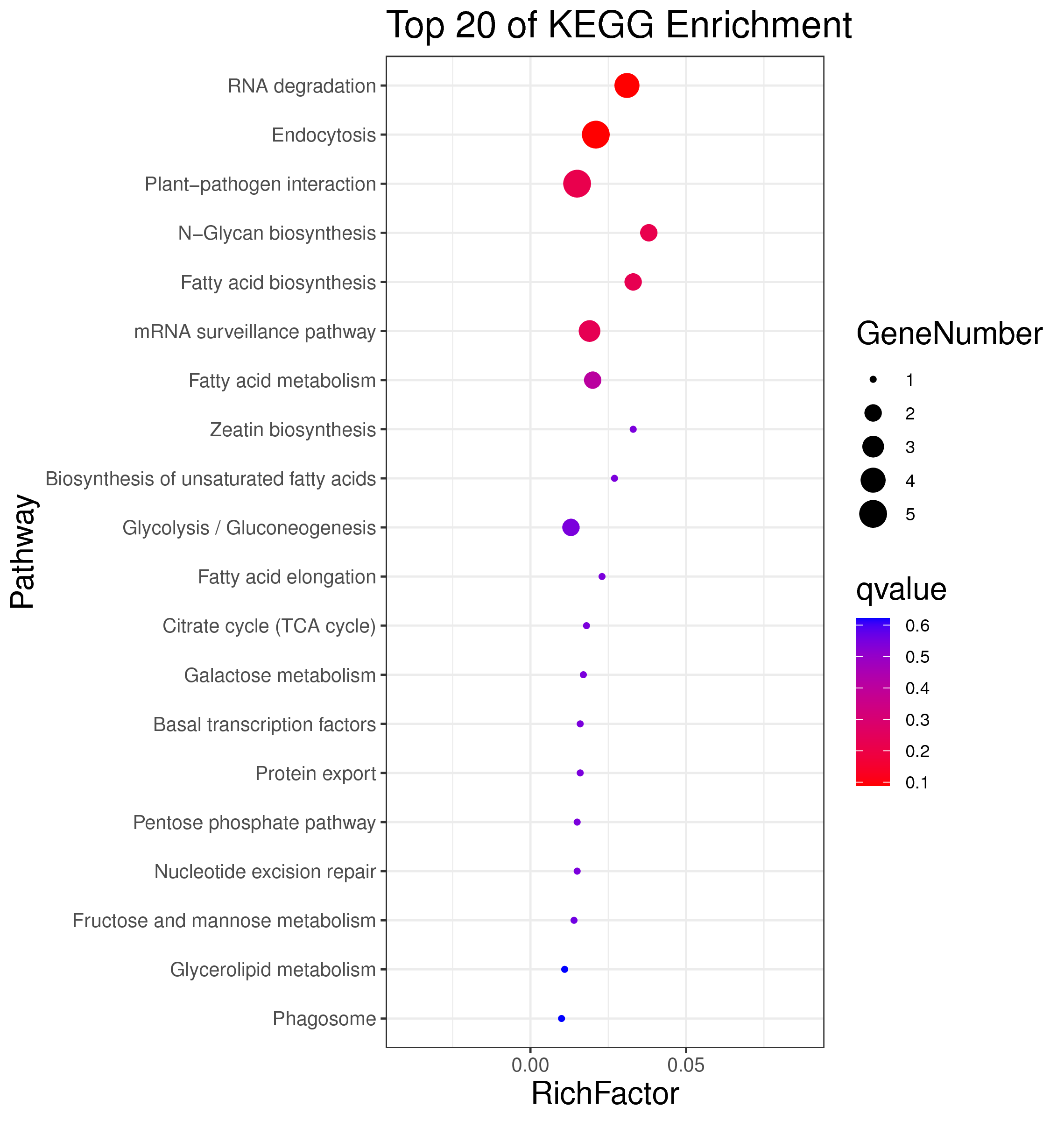
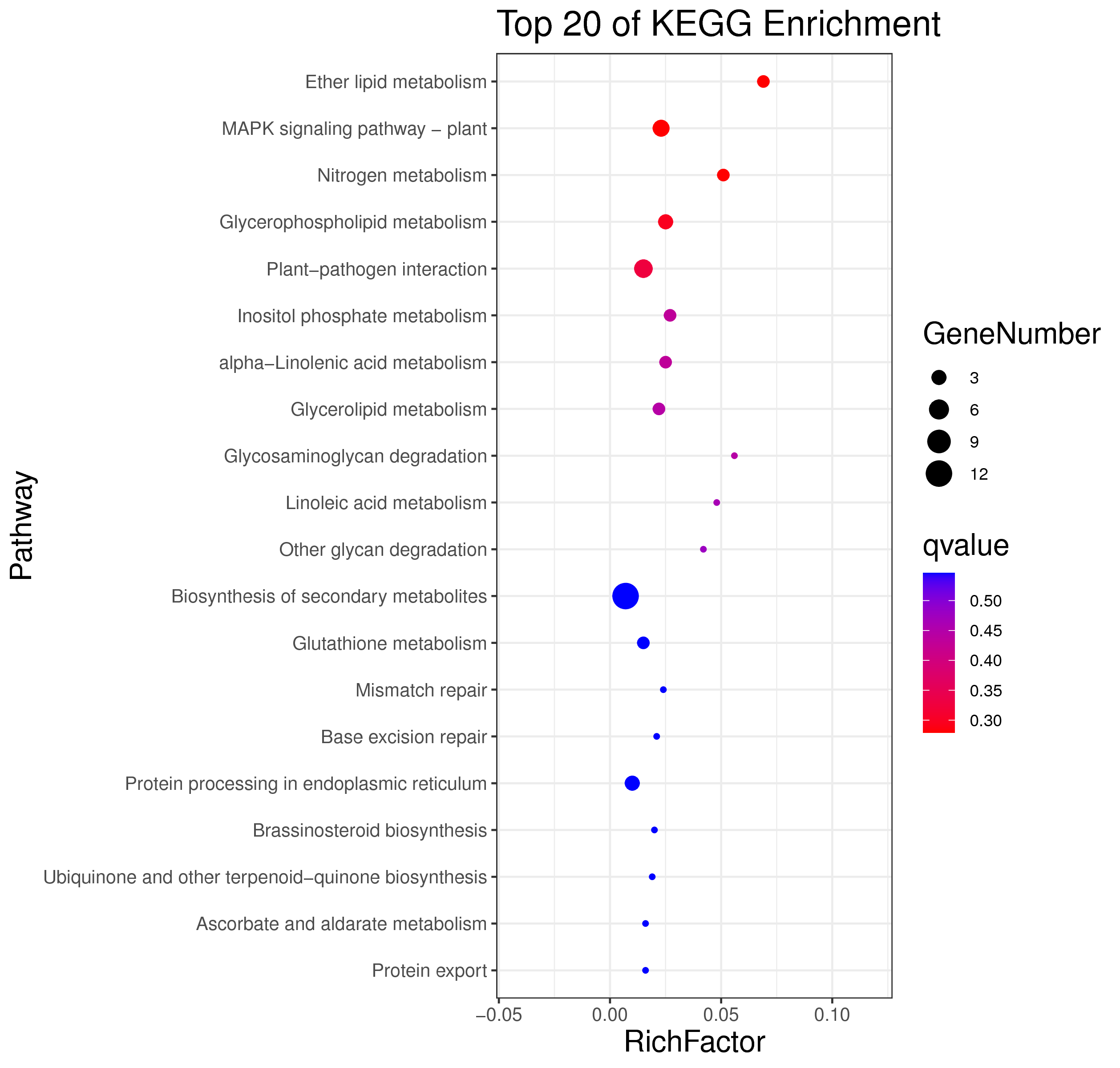
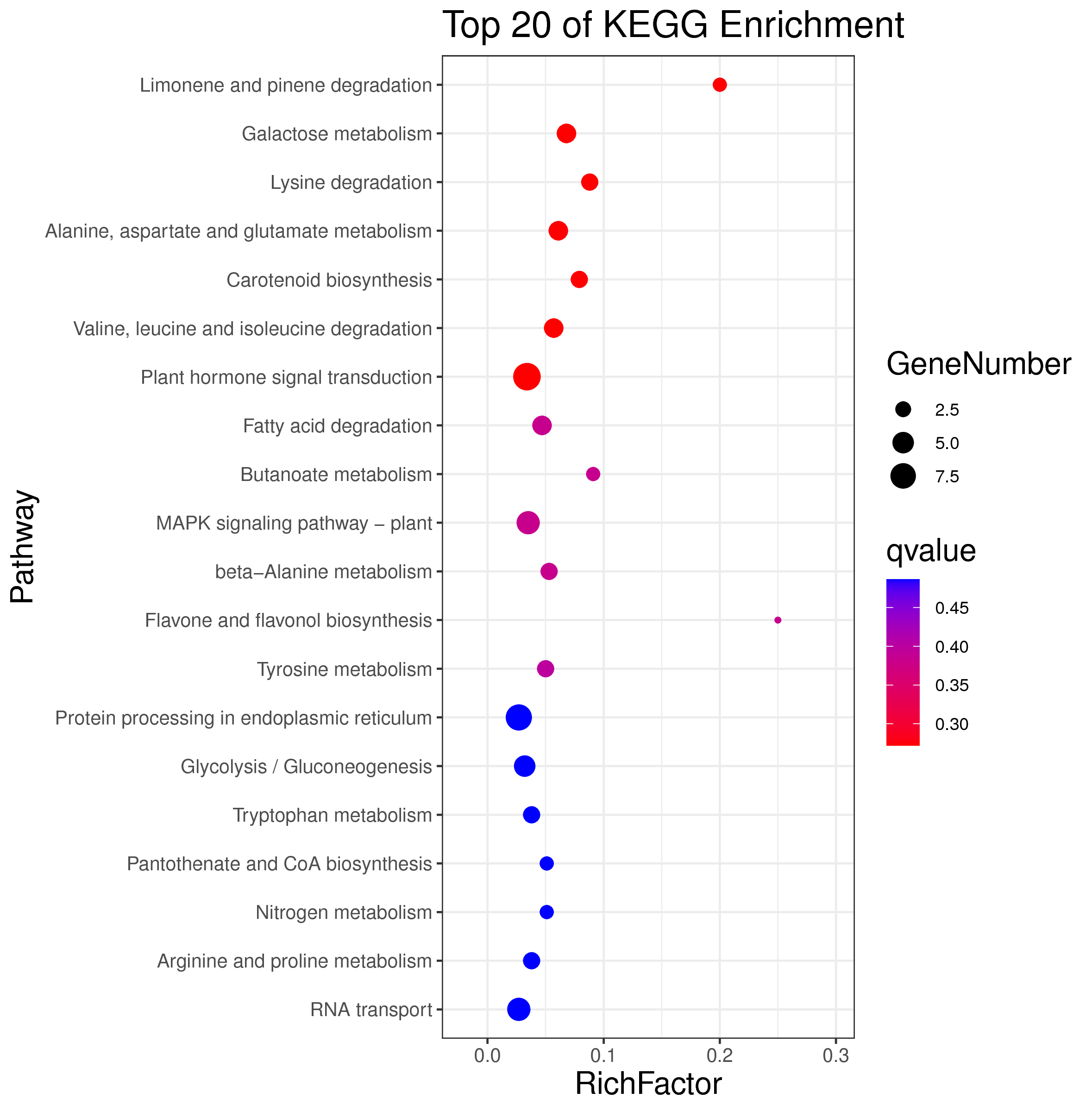
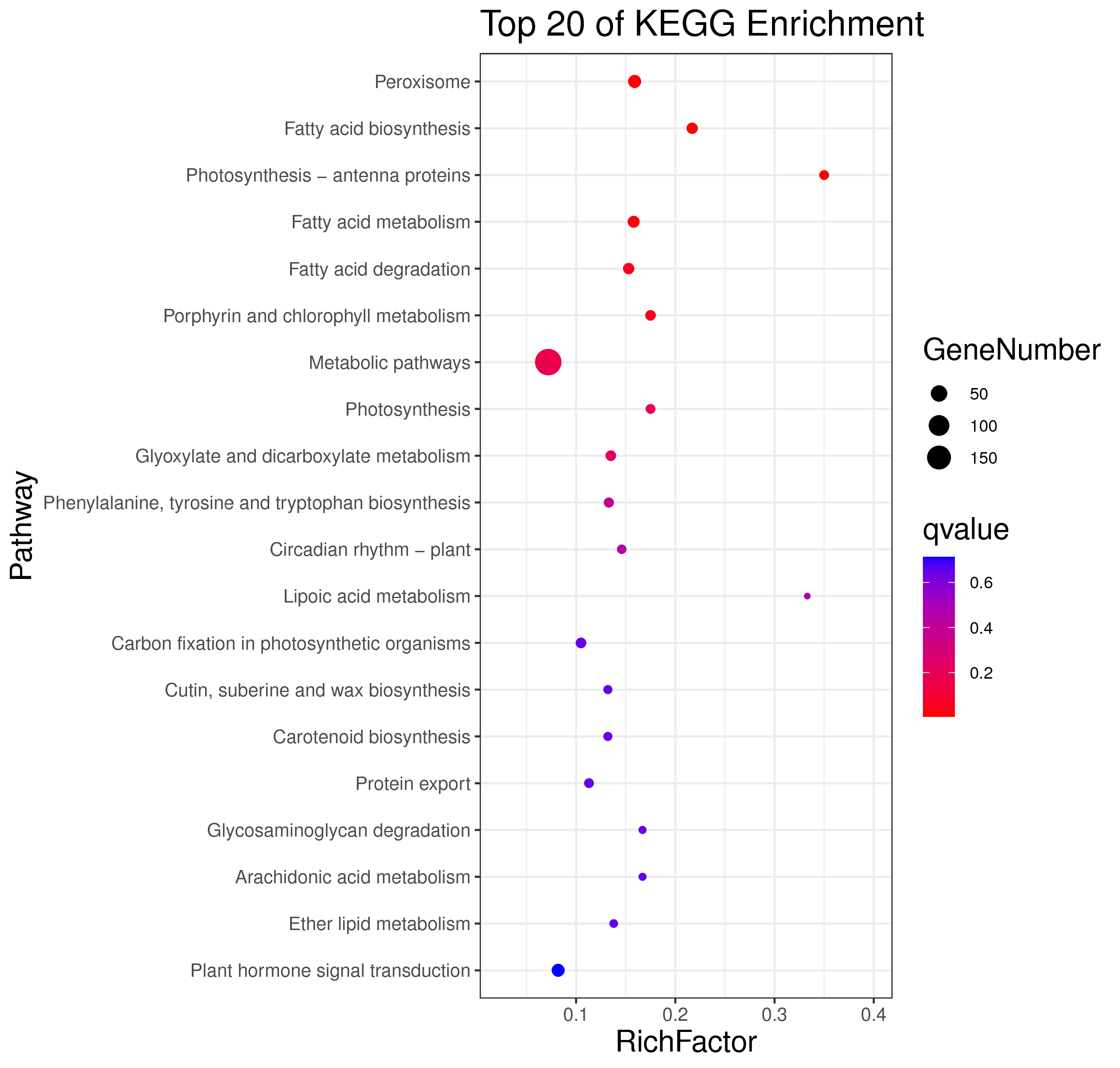
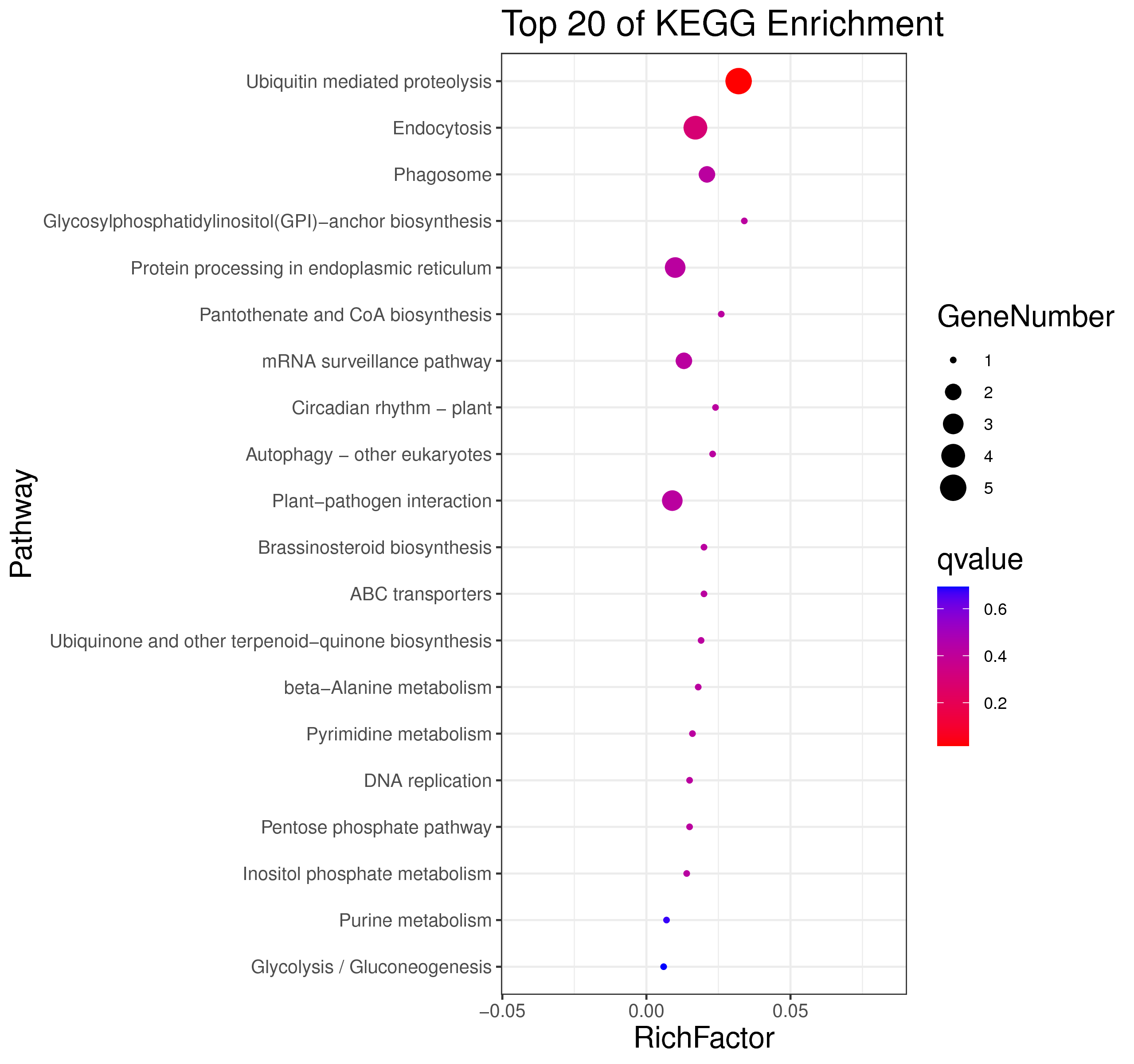
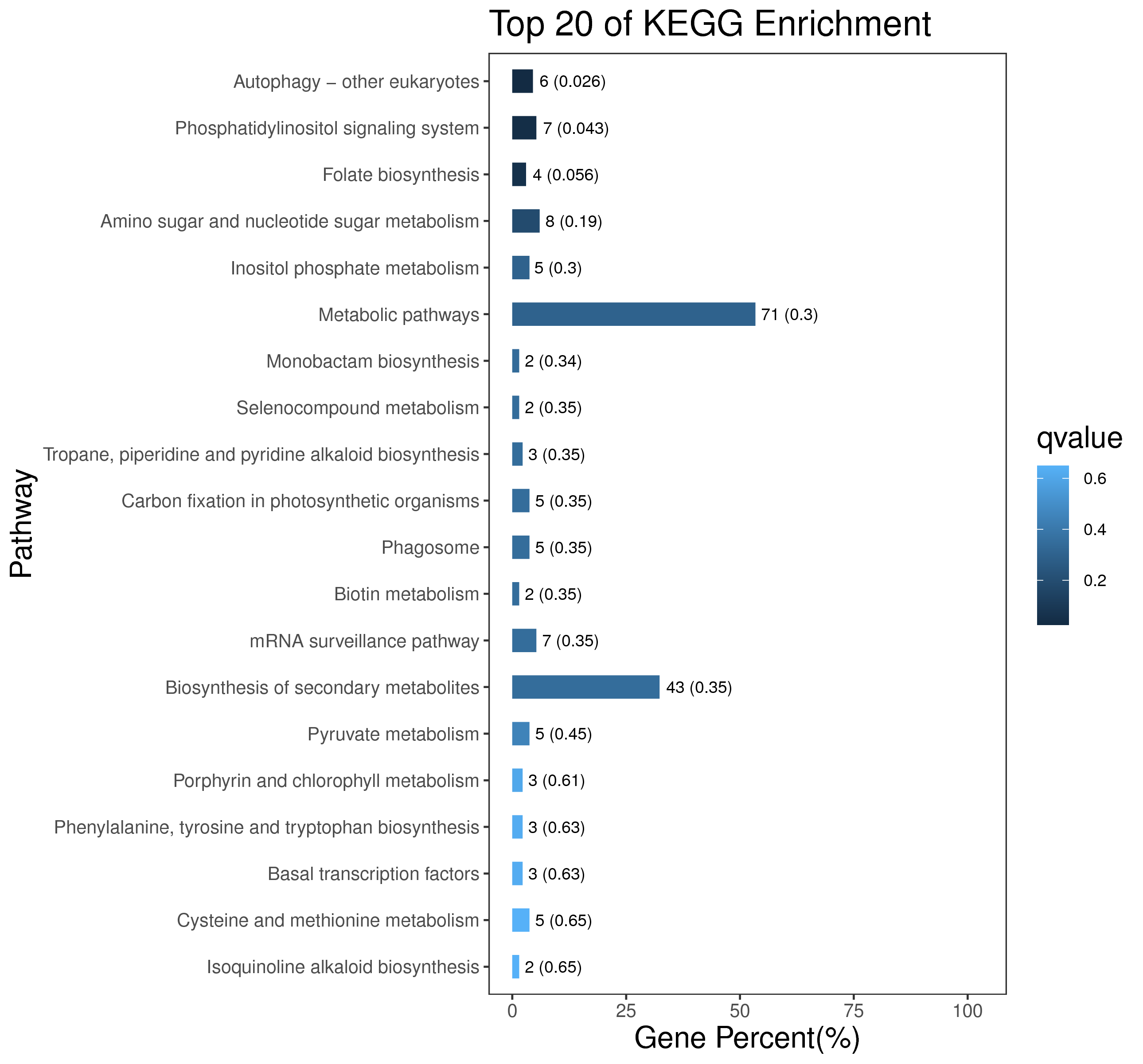
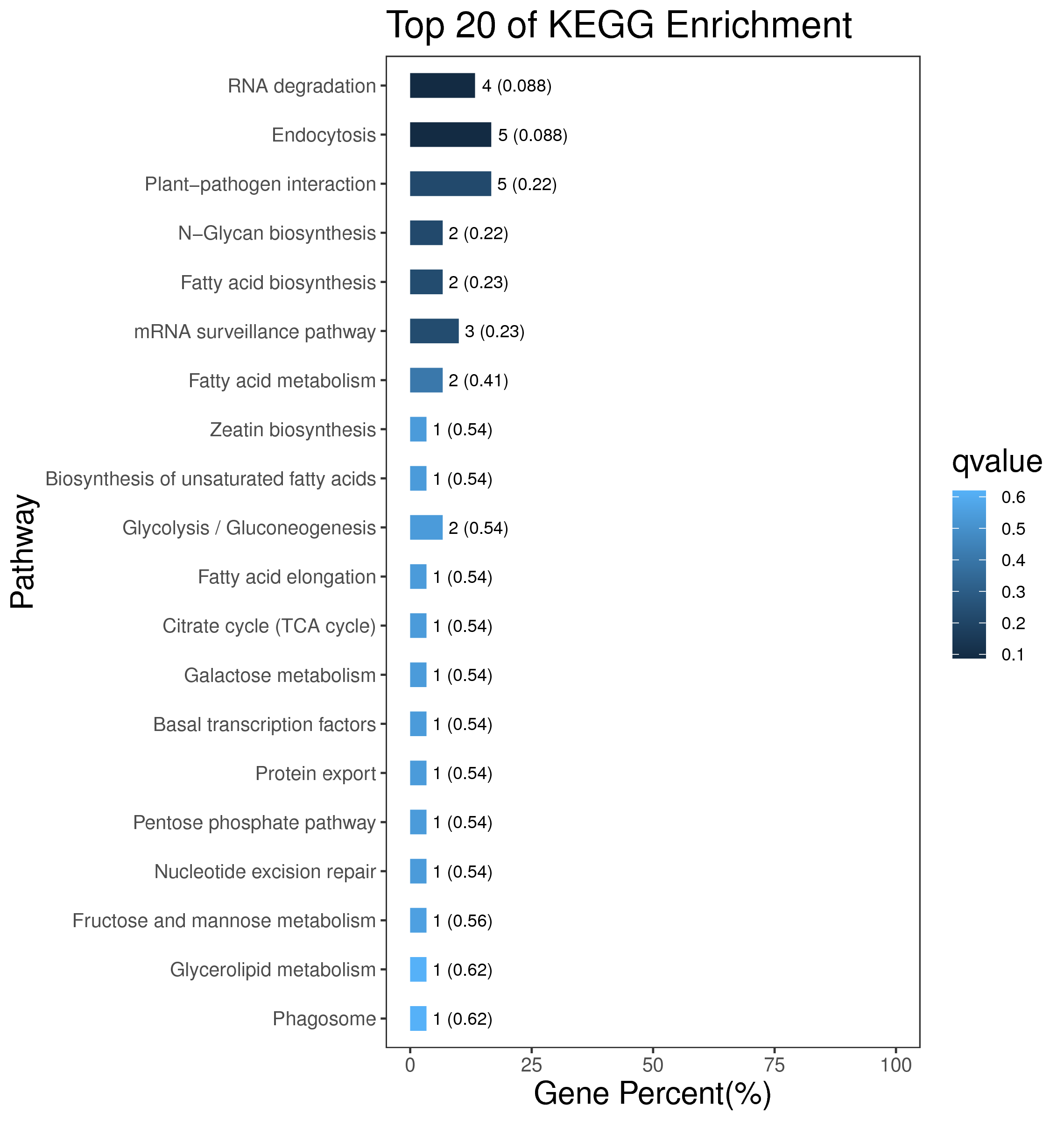
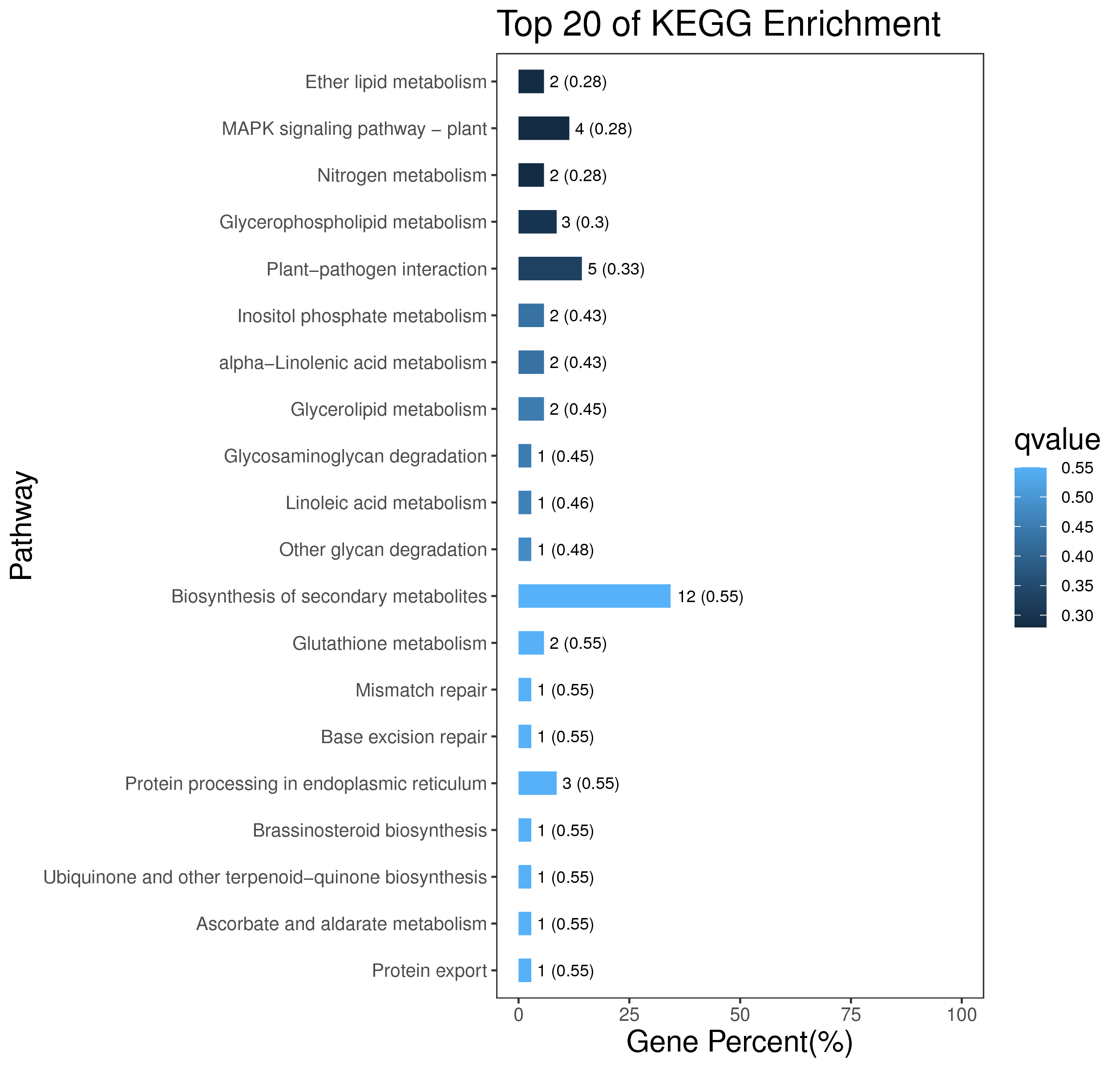
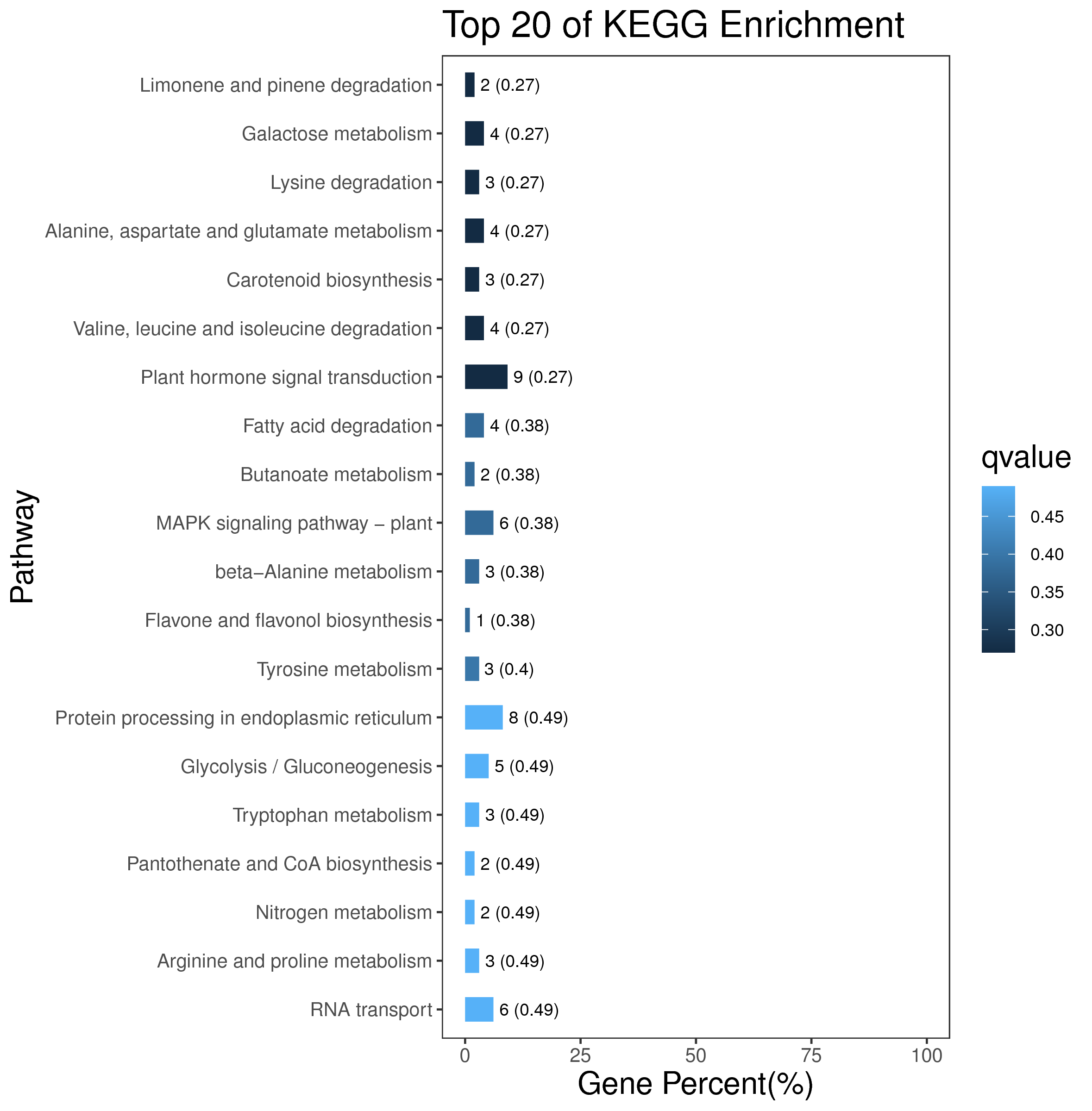
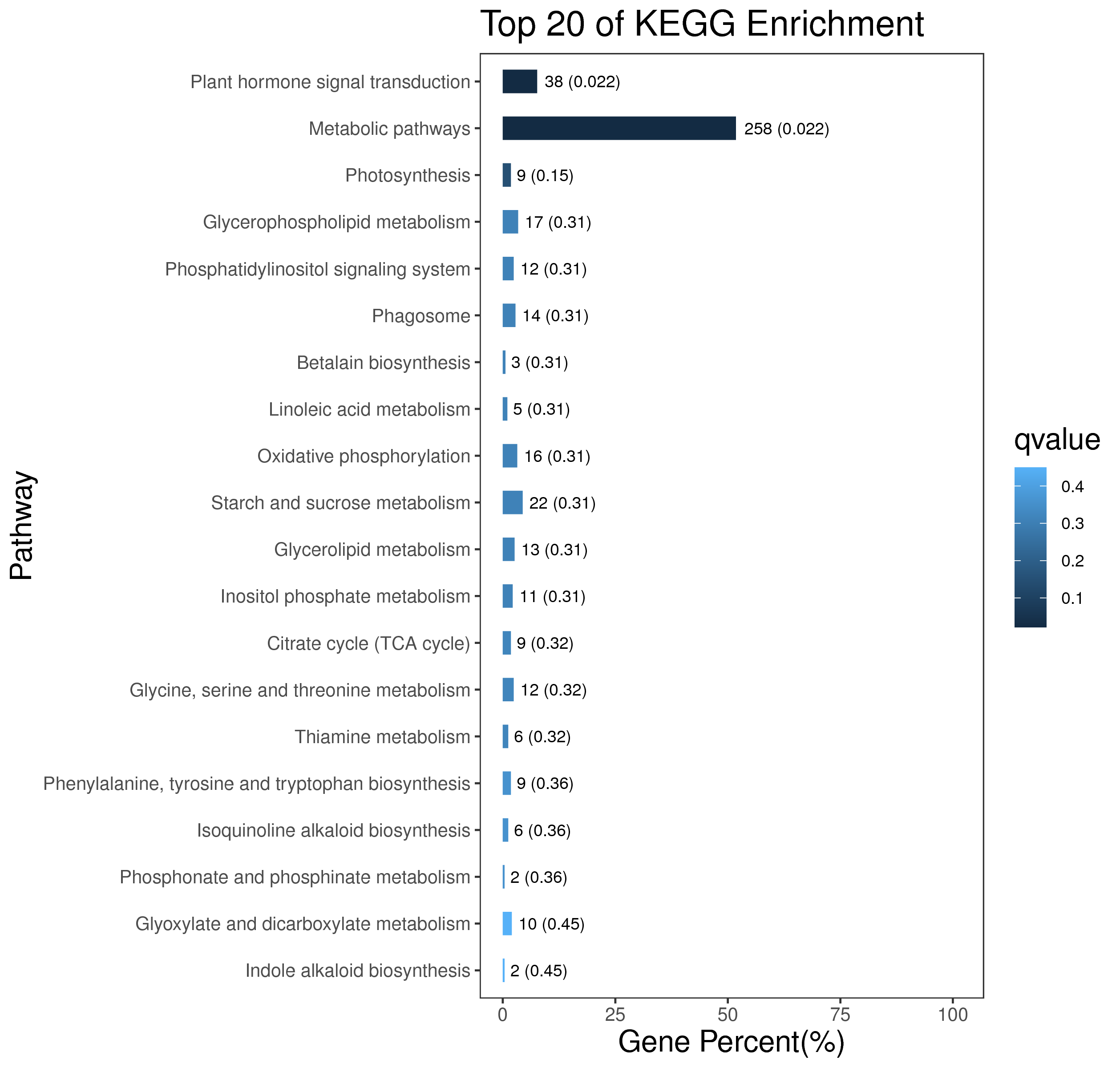
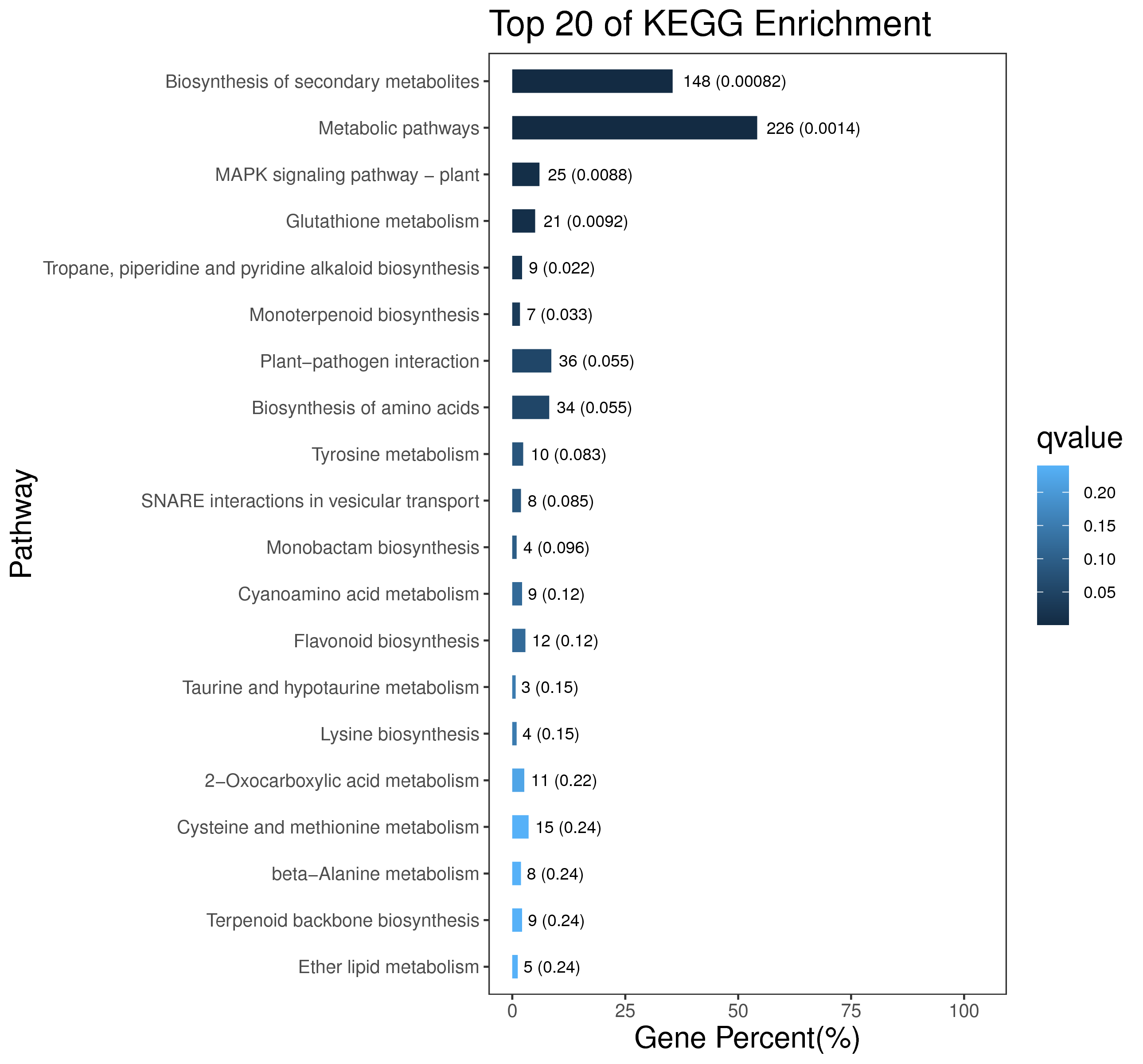
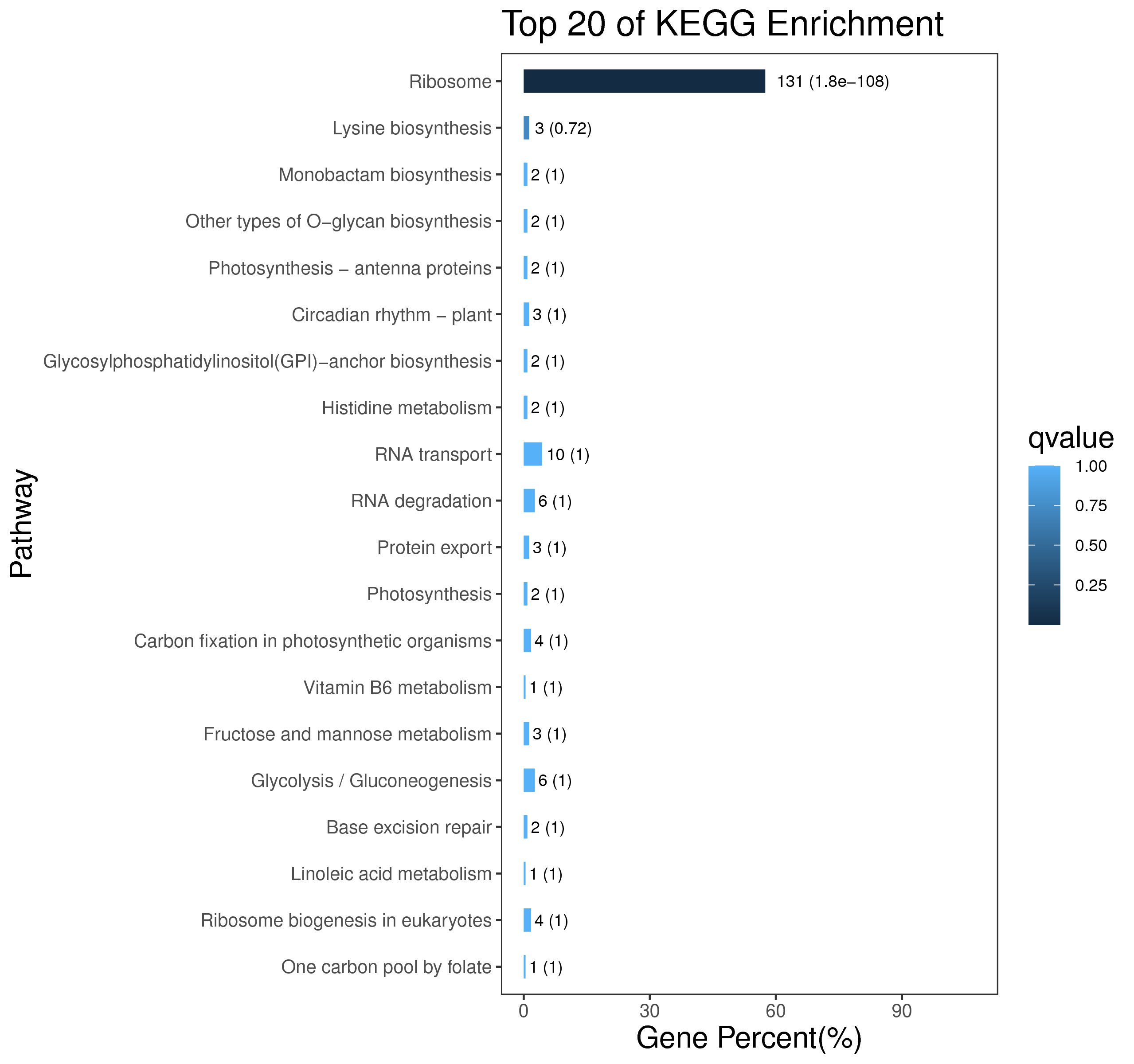
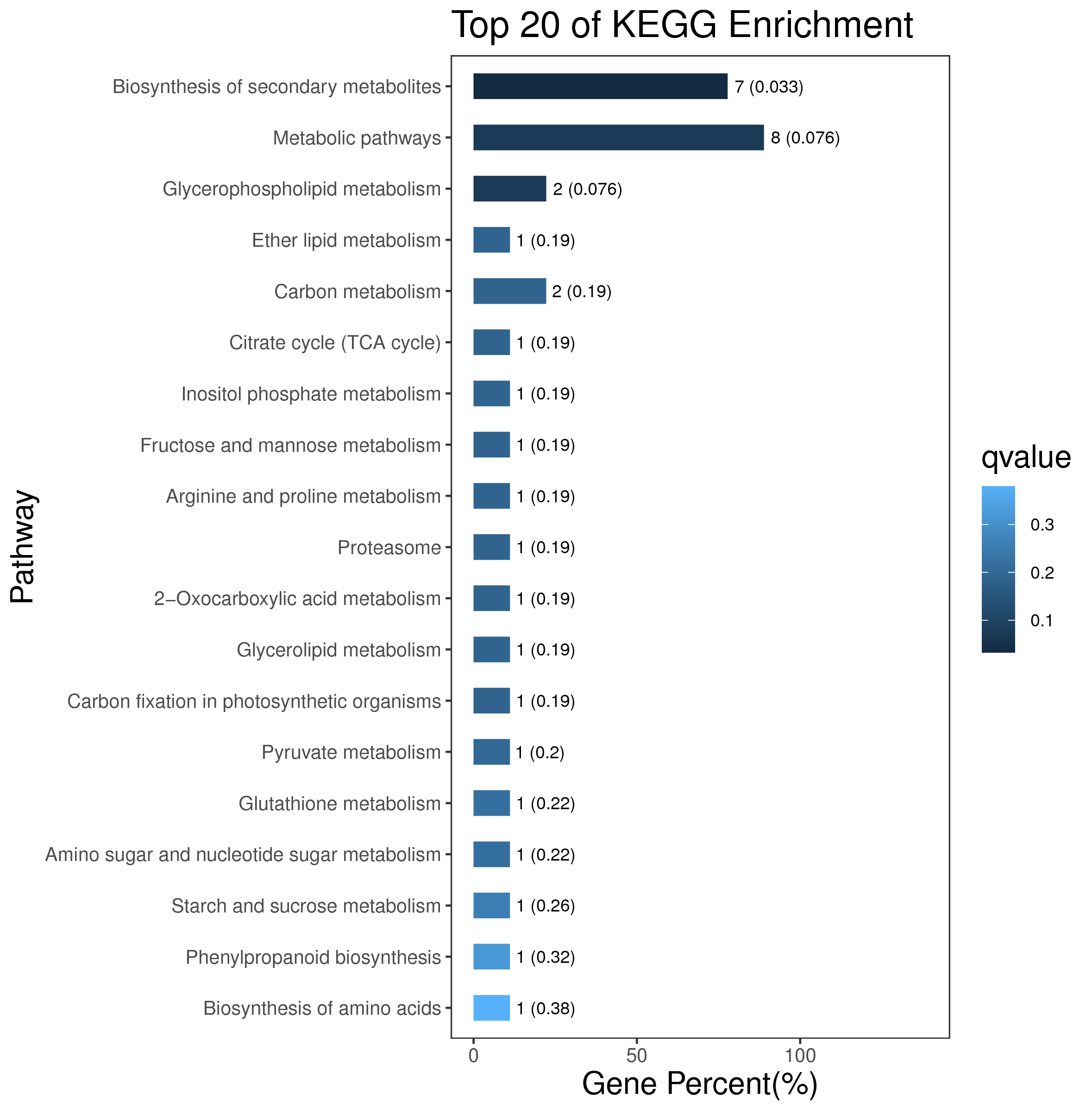
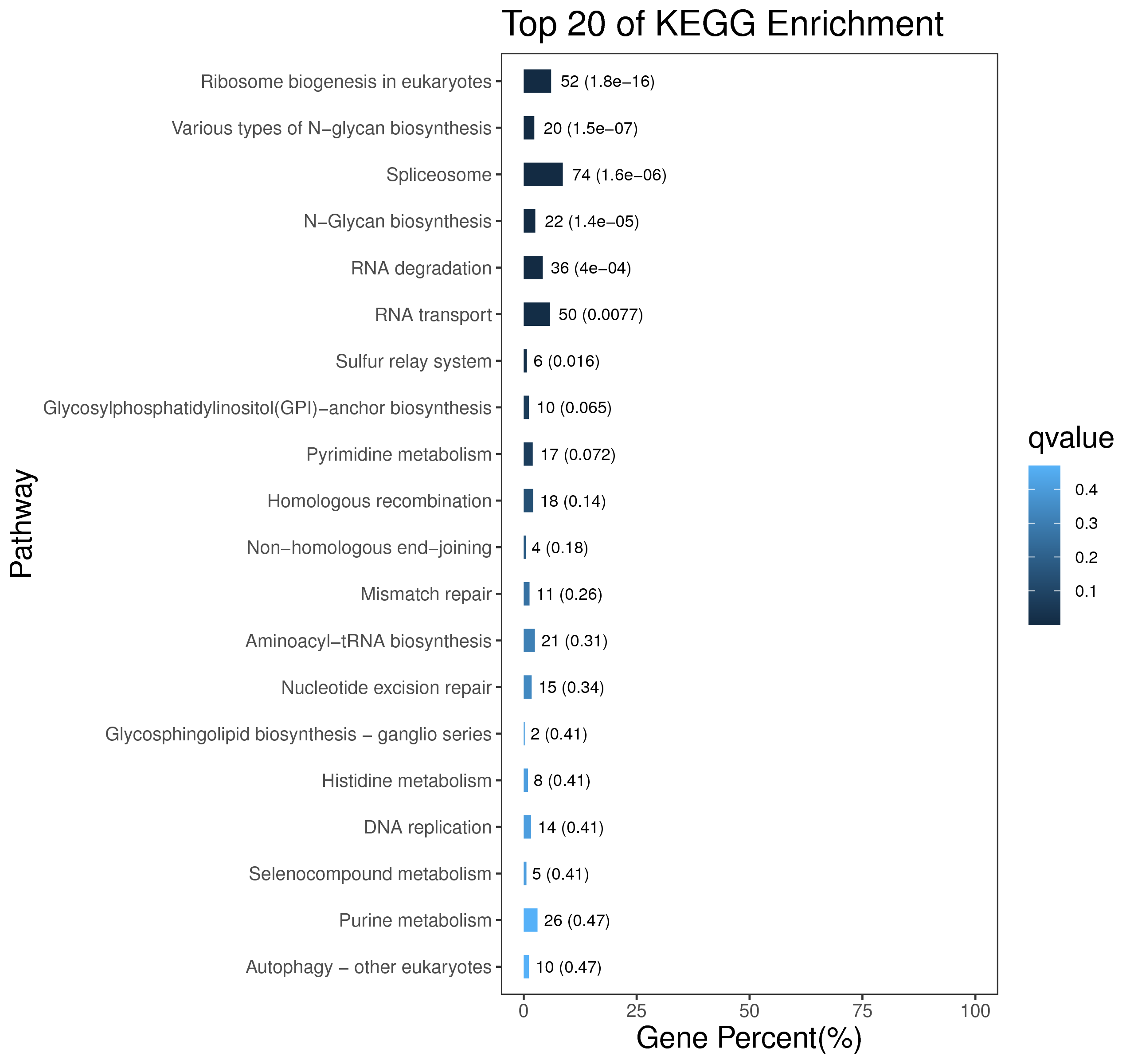
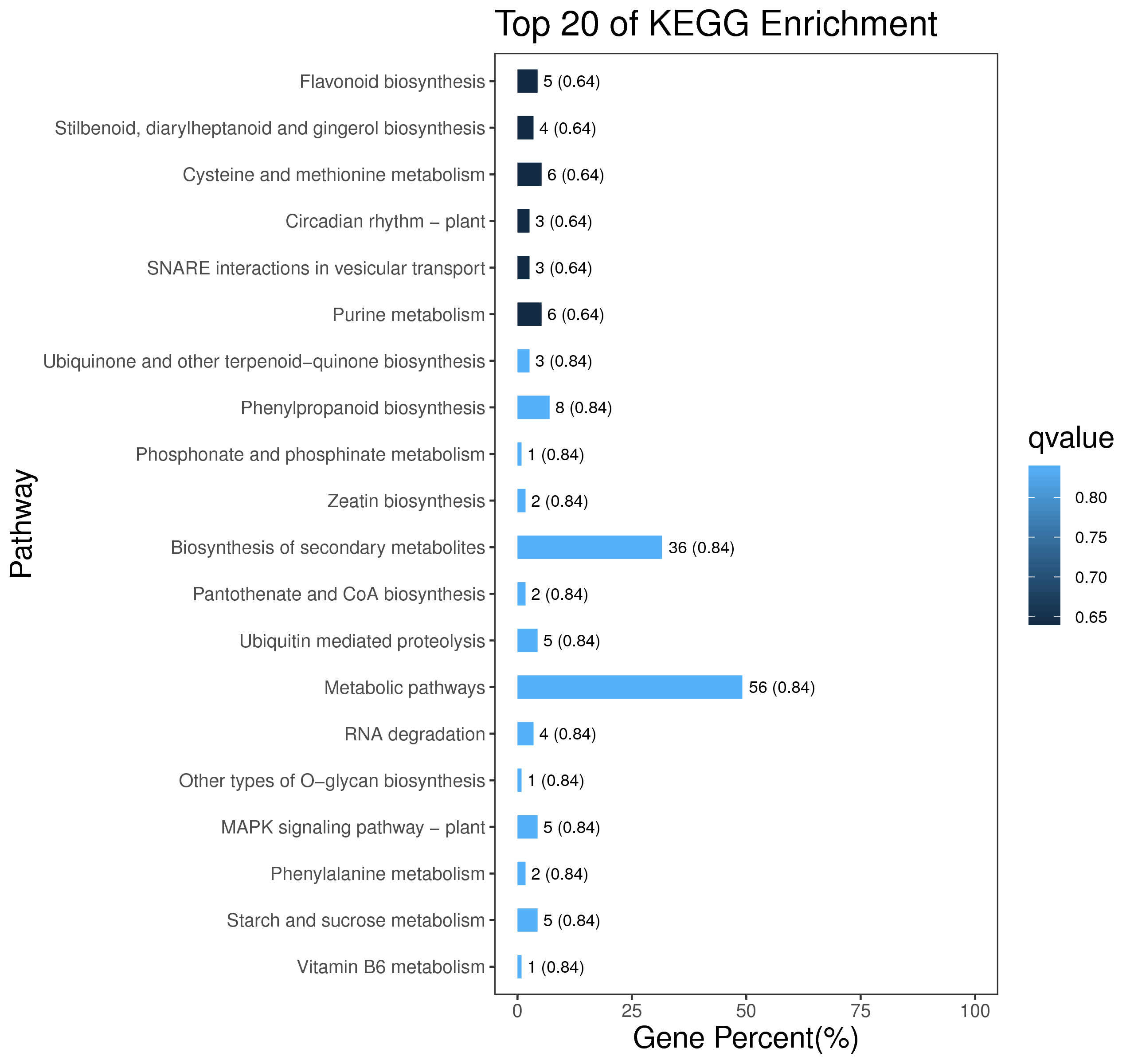
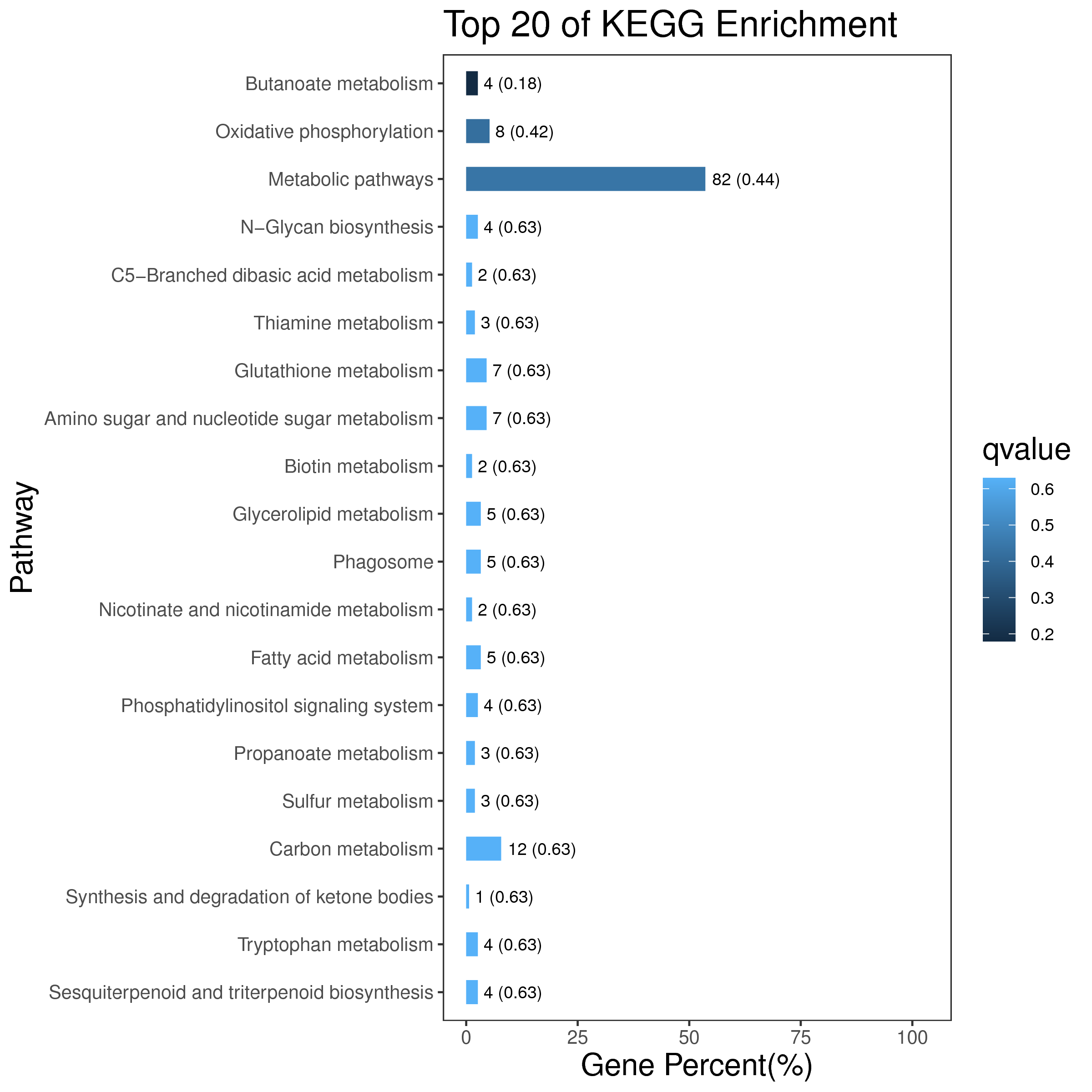
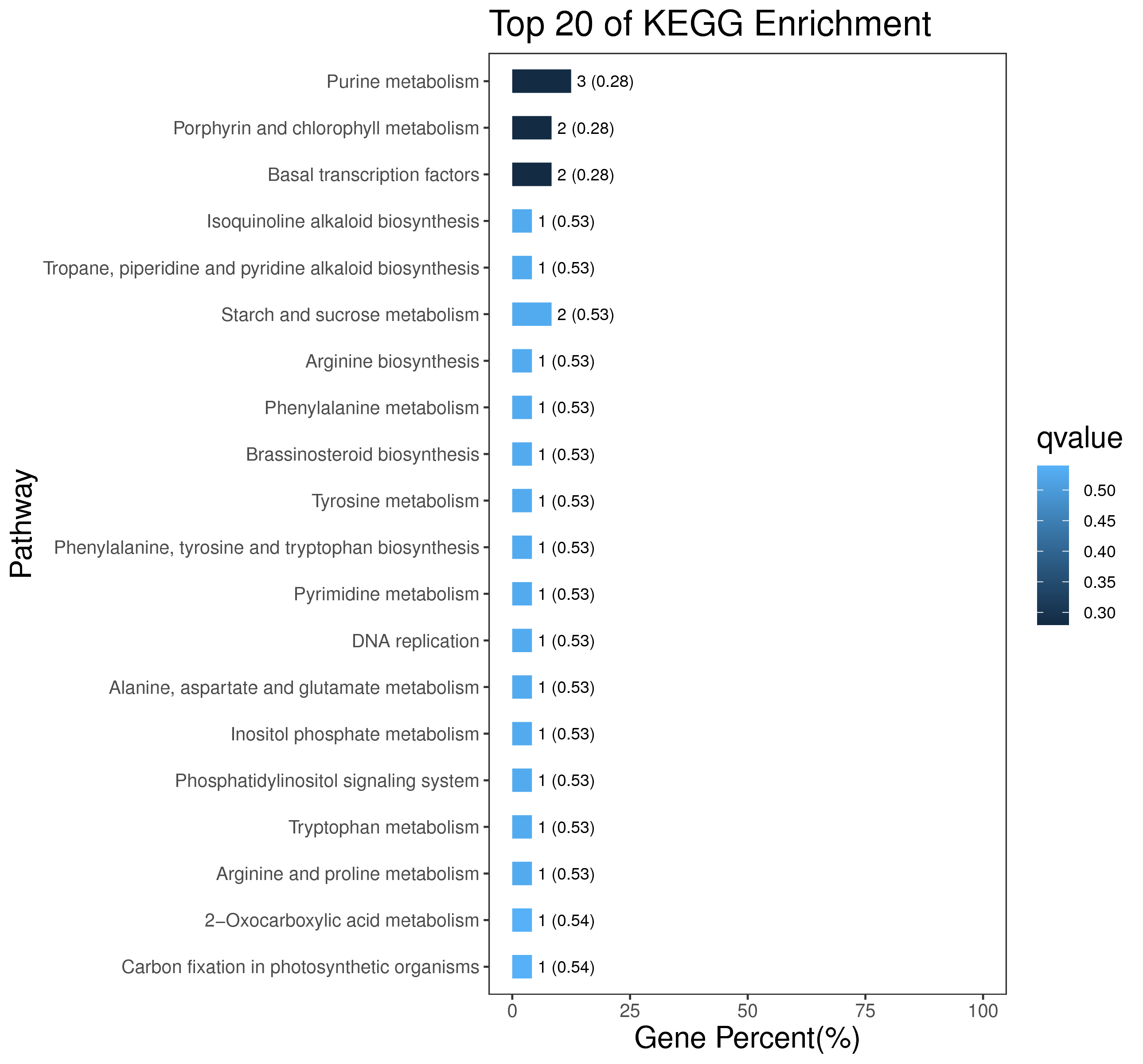
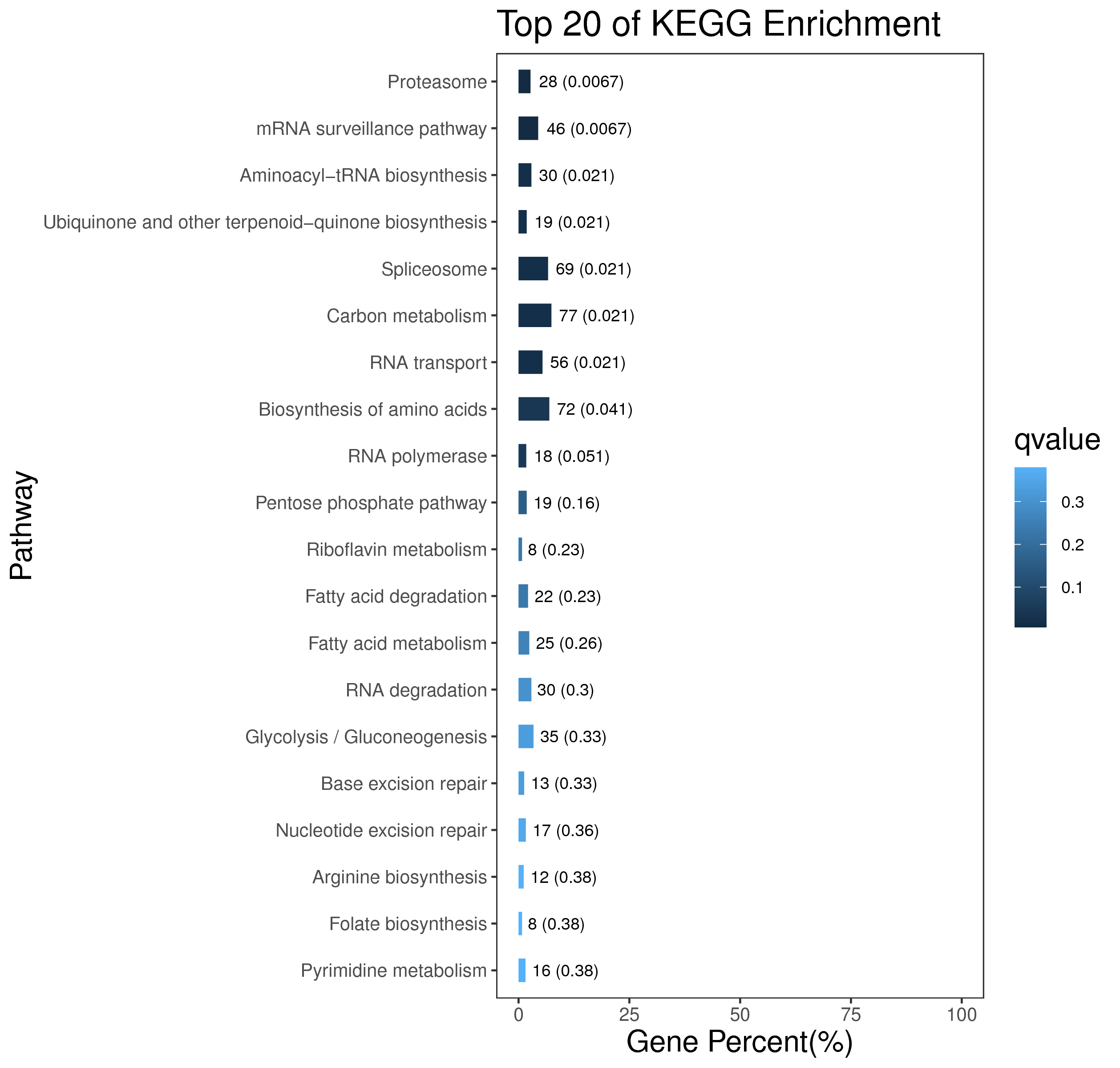
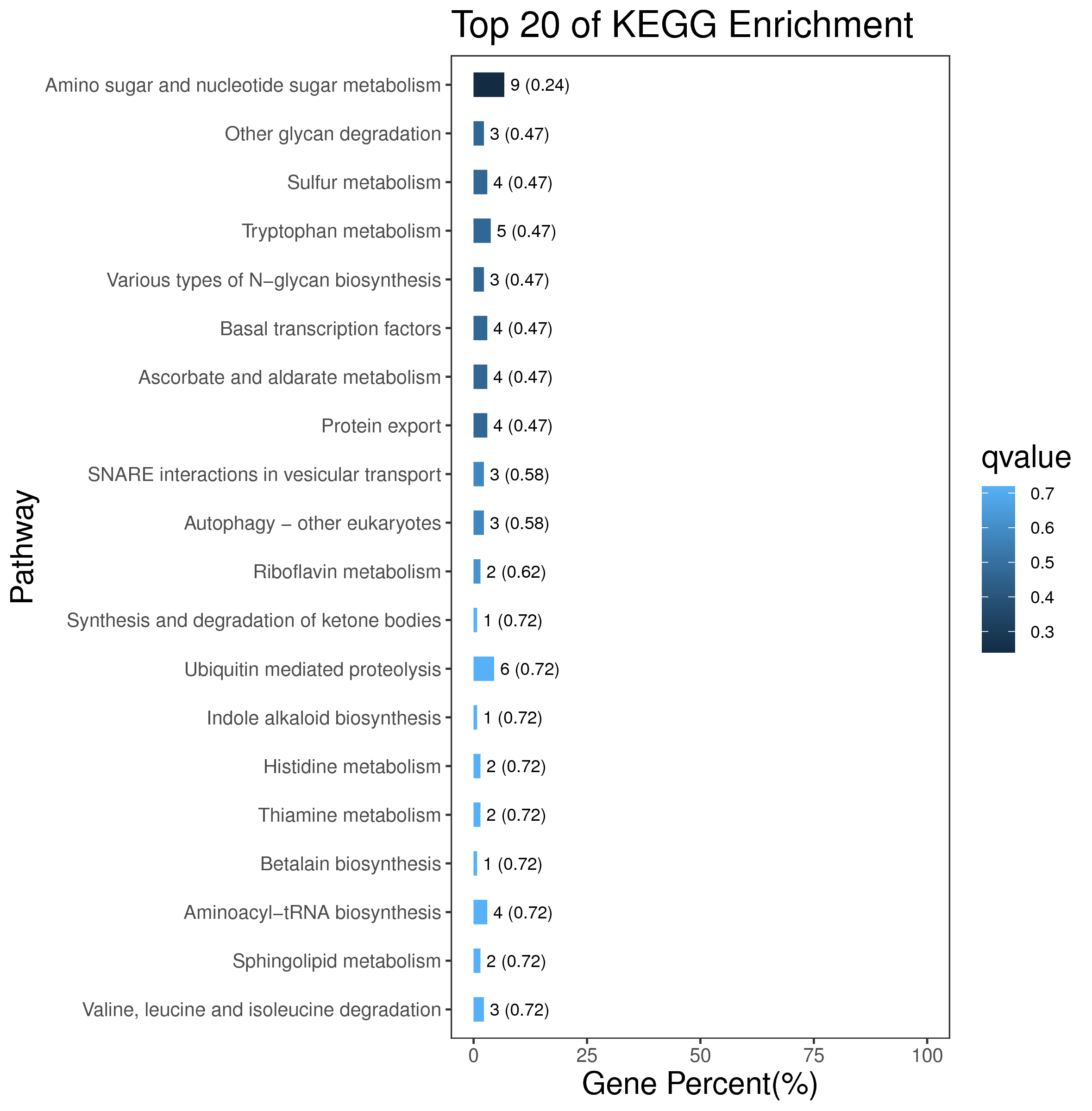
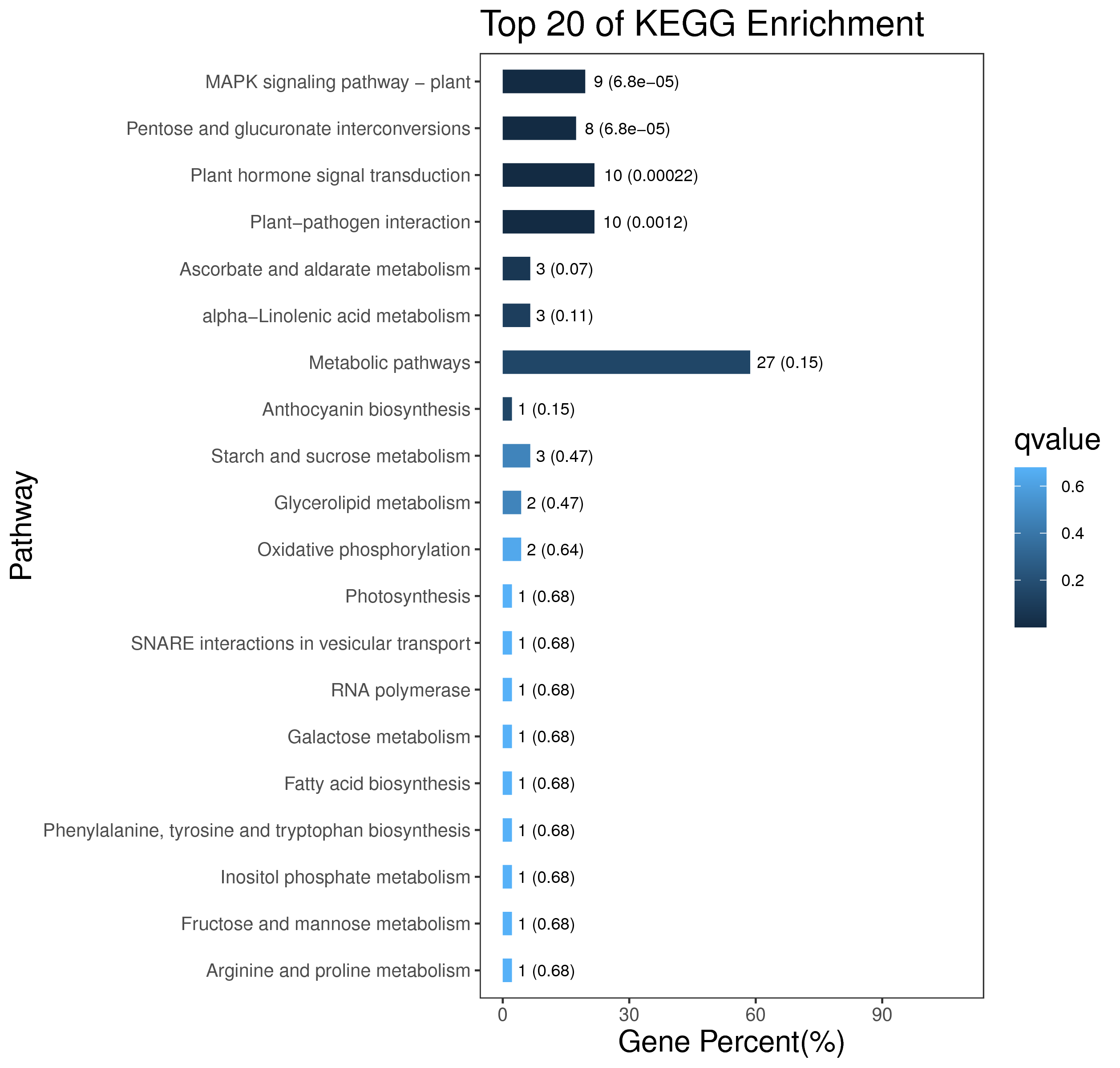
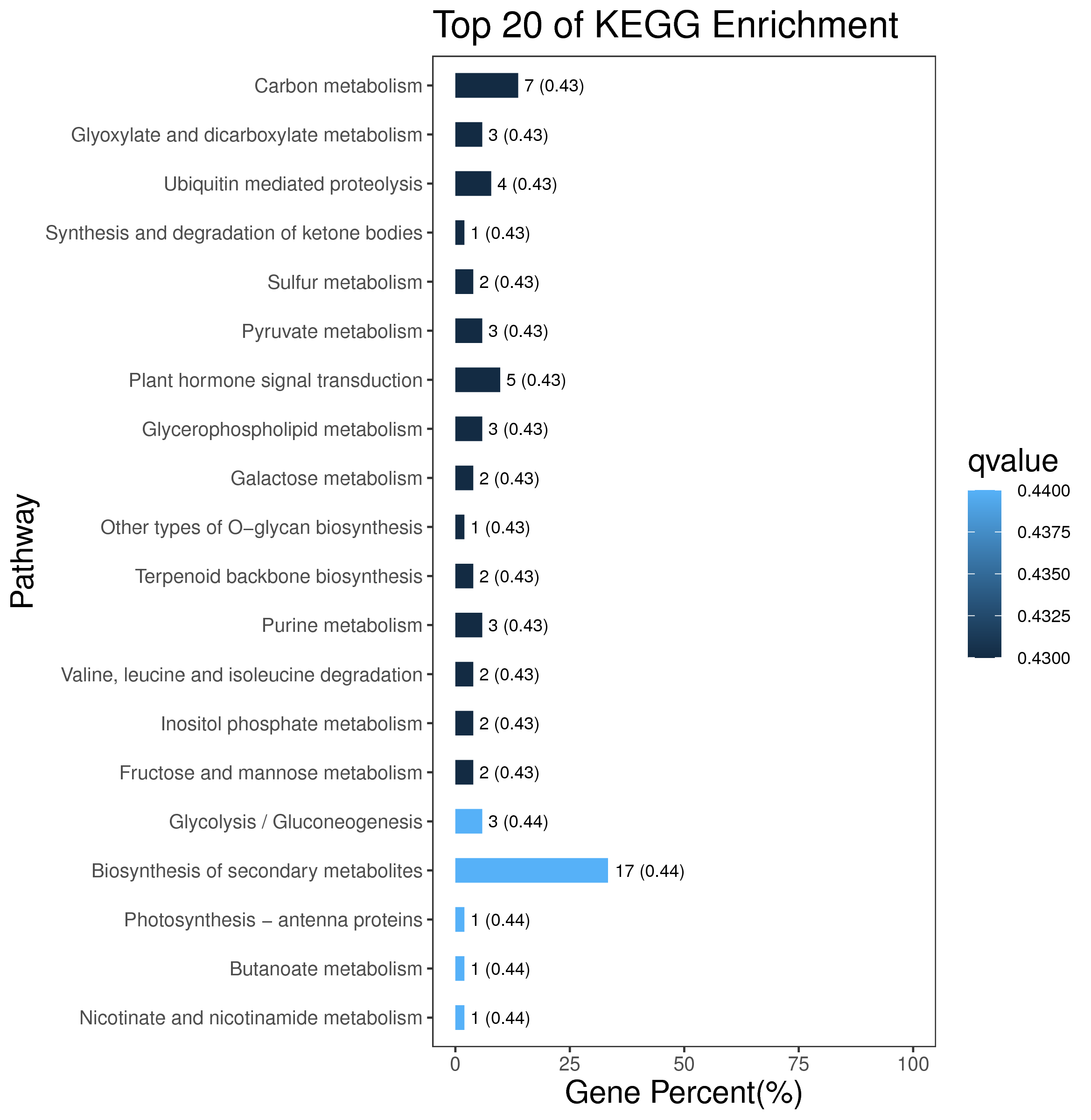
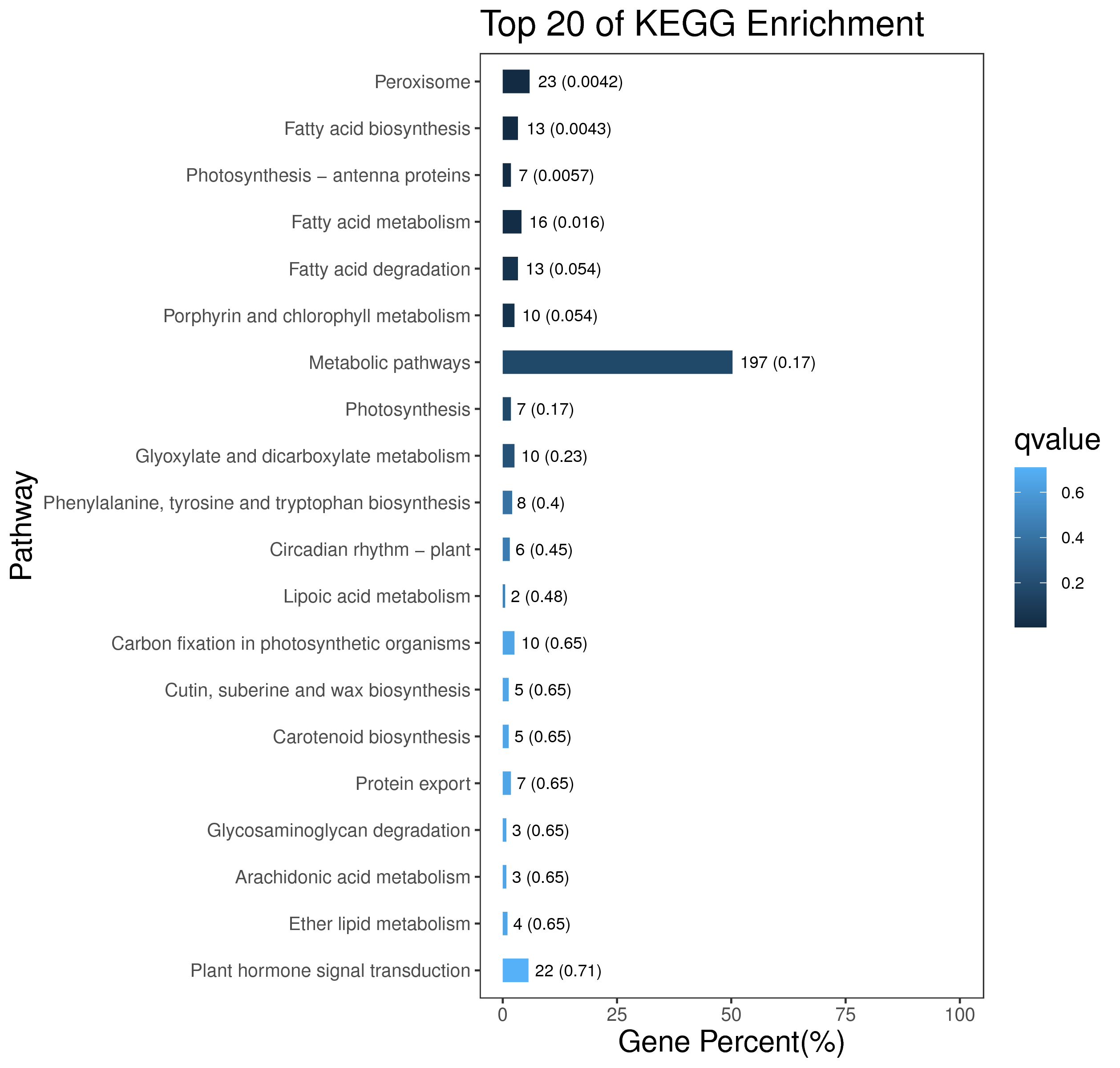
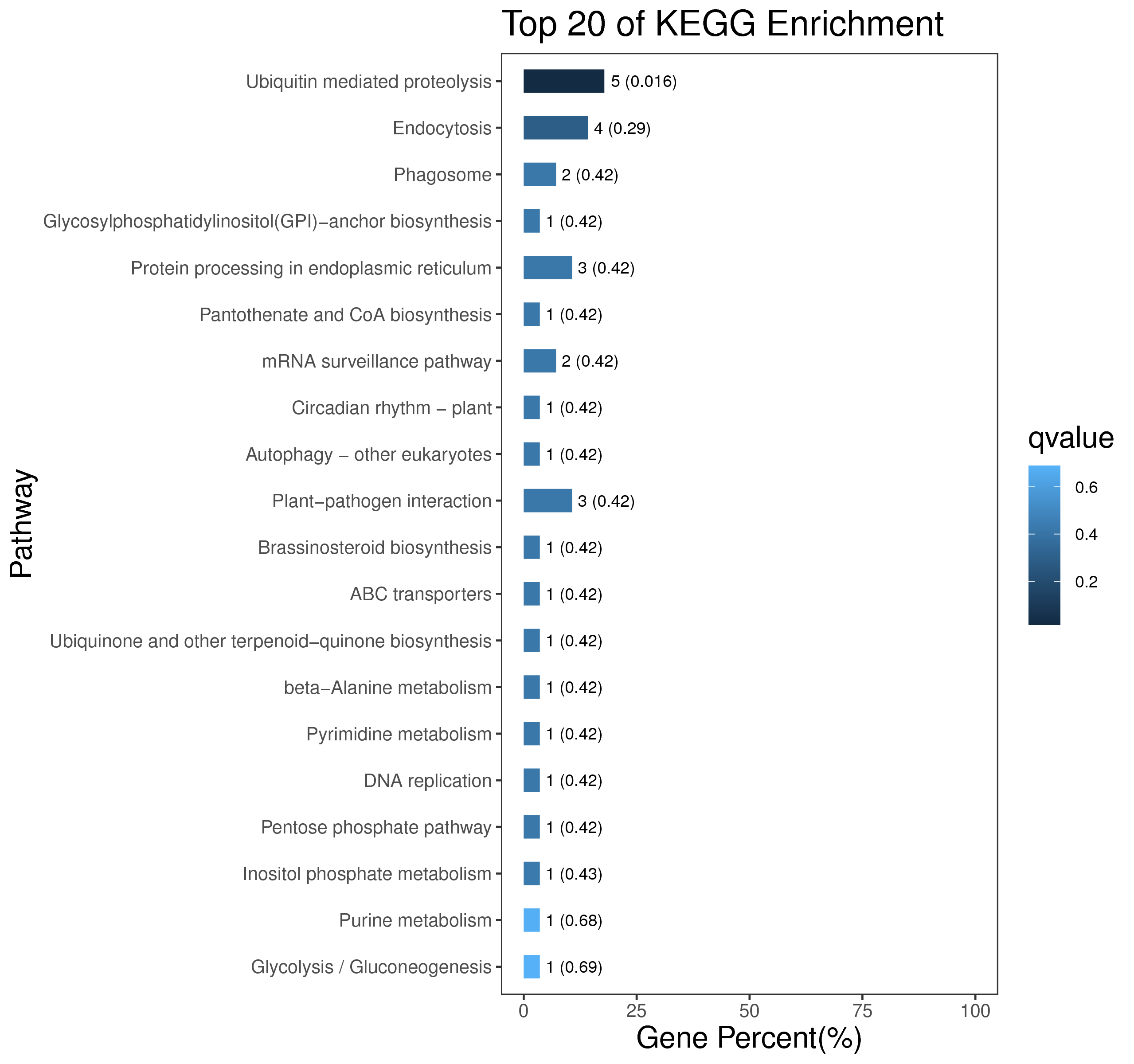
1 项目概述
| 项目编号 | GDMR20030248_sup_18 |
| 项目内容 | Rosa_chinensis_12_WGCNA |
| 参考基因组 | Rosa_chinensis |
2 流程介绍
WGCNA[1](weighted gene co-expression network analysis,权重基因共表达网络分析)是一种分析多个样本基因表达模式的分析方法,可将表达模式相似的基因进行聚类,并分析模块与特定性状或表型之间的关联关系,因此在疾病以及其他性状与基因关联分析等方面的研究中被广泛应用。
WGCNA算法是构建基因共表达网络的常用算法,使用R语言包[2]进行分析。WGCNA算法首先假定基因网络服从无尺度分布,并定义基因共表达相关矩阵、基因网络形成的邻接函数,然后计算不同节点的相异系数,并据此构建分层聚类树(hierarchical clustering tree),该聚类树的不同分支代表不同的基因模块(module),模块内基因共表达程度高,而分属不同模块的基因共表达程度低。最后,探索模块与特定表型或疾病的关联关系,最终达到鉴定疾病治疗的靶点基因、基因网络的目的。
WGCNA 与趋势分析都为基因共表达分析方法,与趋势分析相比,WGCNA 有以下优势:
1.聚类方法:使用权重共表达策略(无尺度分布),更加符合生物学现象;
2.基因间的关系:能呈现基因间的相互作用关系,且能找出处于调控网络中心的 核心基因(hub gene);
3.样本数:适合于大样本量,且越多越好。而趋势分析 5 个点以上就会使结果非常复杂,准确性 下降,且最多只能分析 8 个点;
4.与表型的关联:可以利用模块特征值和 hub 基因与特定性状、表型进行关联分析,更准确地分析生物学问题。
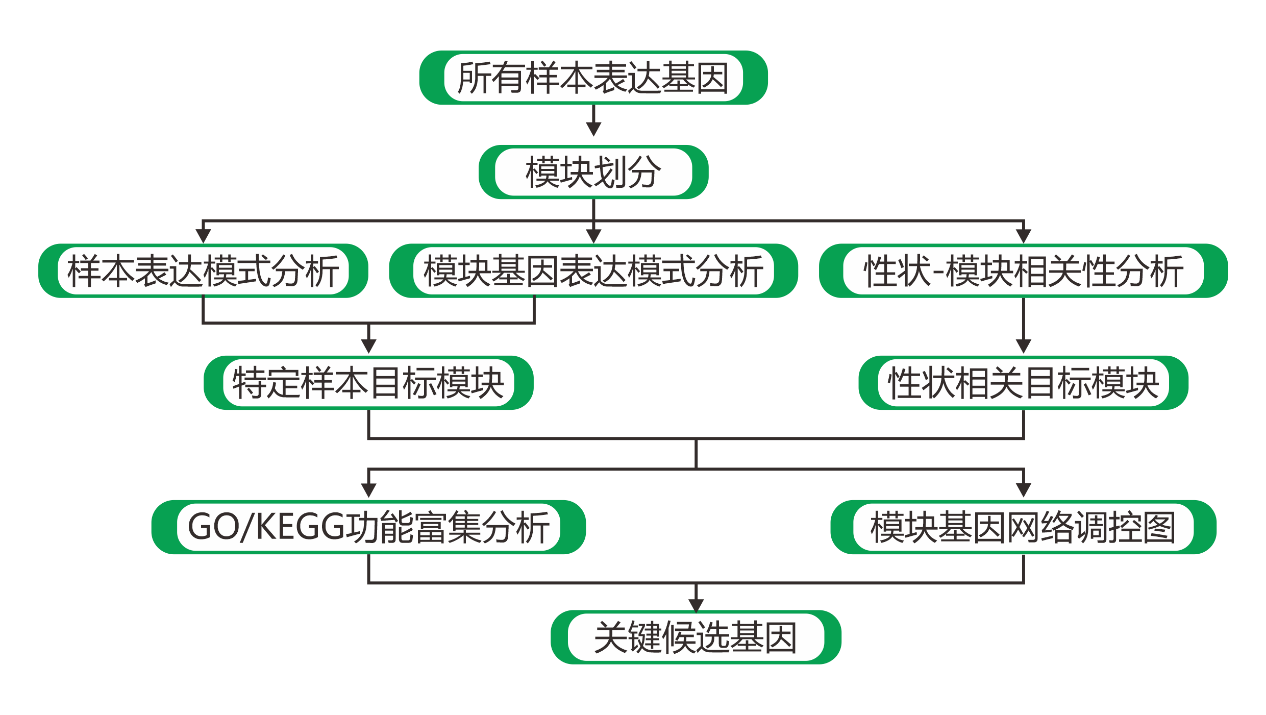 |
| Fig 2-0-1 WGCNA总体分析流程图 |
3 数据过滤
在进行WGCNA分析之前,我们需要对选用的基因集进行筛选过滤,把低质量的对结果造成不稳定影响的基因或样品从中去掉,提高网络构建的精度。
4 模块划分
基因间具有相互诱导、阻遏表达或协同作用,这些作用都会导致相关基因的表达量间存在相关性,在大样本的情况下,基因的表达分类更加有规律,利用WGCNA分析可以将具有相似表达模式的基于进行归类,将成千上万的基因划分成数个至数十个的模块。基于模块的分析更利于对相似基因进行分析,并且结合表型信息能更好的找出与特定性状相关联的模块。
4.1 样本层次聚类树
首先我们根据所有基因的表达量对所有样本进程层次聚类,将每个样本视为一个簇,计算各个簇之间的距离,最近的两个簇聚合成一个新的簇,重复以上过程直至最后只有一个簇。通过层次聚类可以查看是否存在离群样品,以便于分析样本情况。
下图所示,关系越相近的样本越容易被归为一个簇,若存在离群样本,则无法归为任何一个簇。
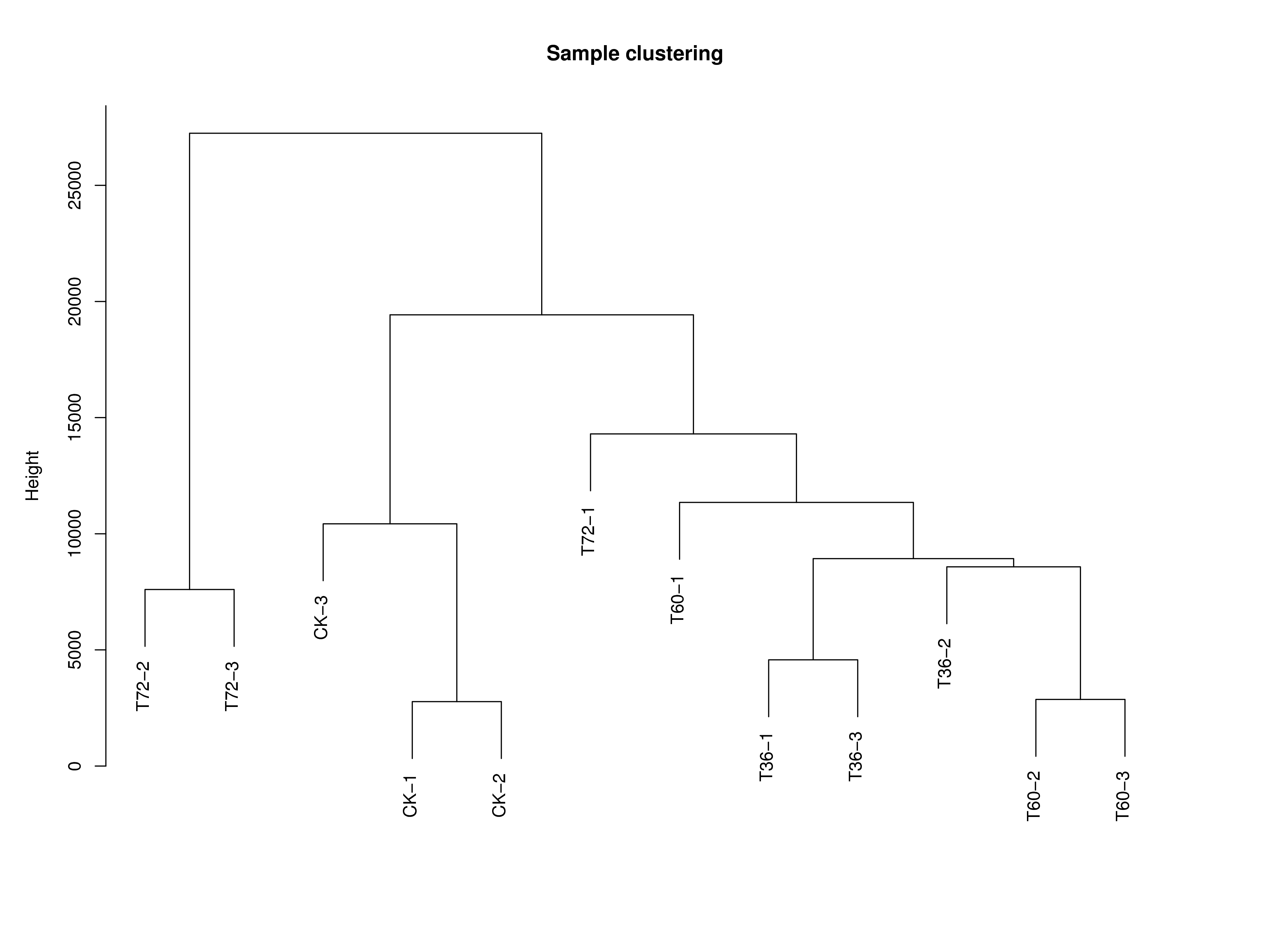 |
| Fig 4-1-1 样本层次聚类树 |
4.2 Power值曲线
对于power值的选取,一般情况下,我们会取相关系数达到平台期(或大于0.8)时最小的Power值作为后续分析的参数(如图4-2-1左图),同时我们会统计在不同power值下,基因平均连通性的变化(如图4-2-1右图)。有时即使选择较大的power值也无法使相关系数大于0.8,可能是由于部分样本与其他样本差别太大,可以通过样本聚类图查看分组信息,以确认有无异常样本,若样本情况正常,也可取经验值进行后续分析。
本次分析选用的Power值参数为:10
本次分析选用的特征参数如下图所示。左图:横坐标表示power值,纵坐标表示相关系数,蓝色横线表示相关系数0.8,红色横线表示相关系数0.9。右图:横坐标表示power值,纵坐标表示基因的平均连接度。
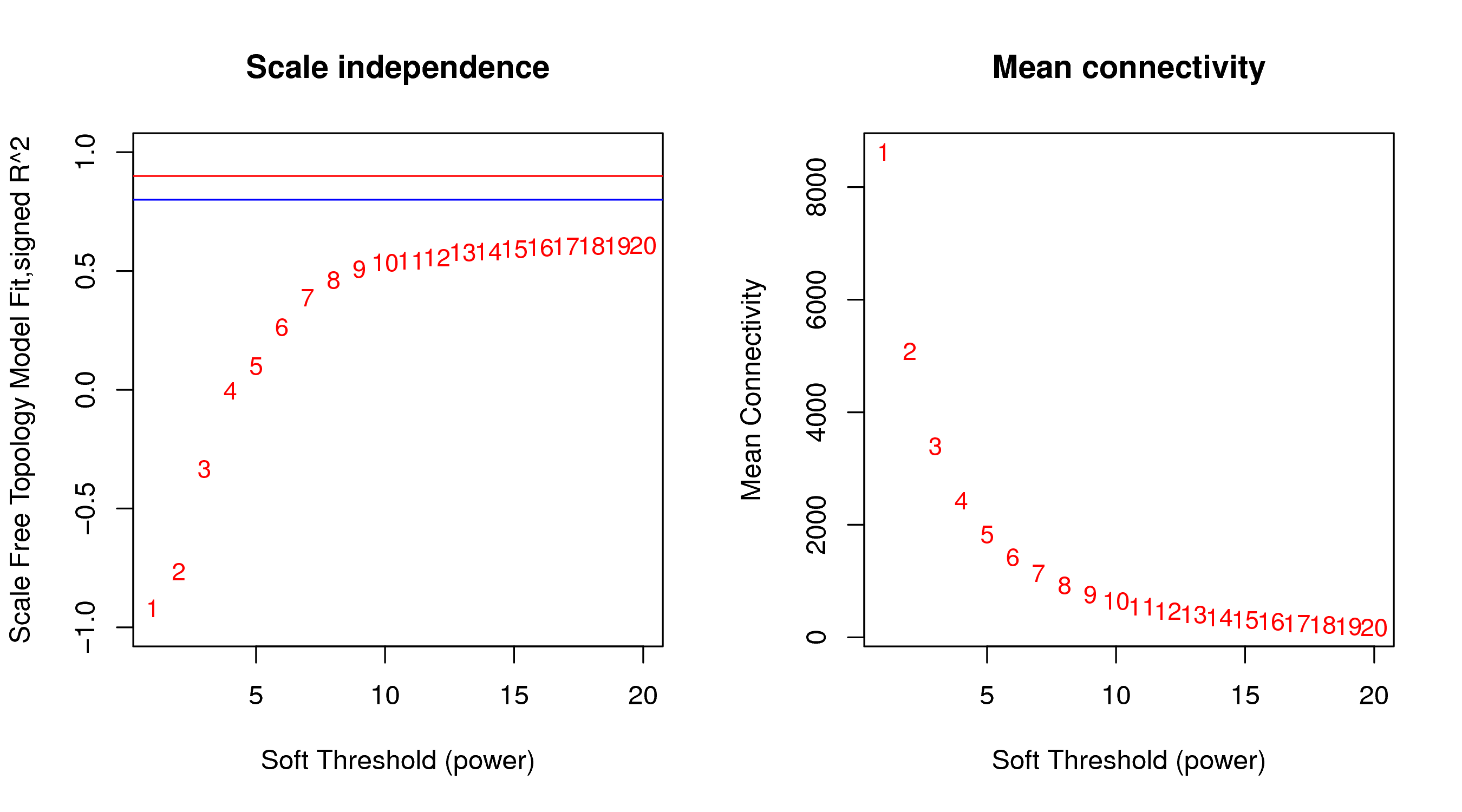 |
| Fig 4-2-1 Power值曲线图 |
4.3 模块划分
根据基因间表达量的相关性构建基因聚类树,并根据基因间的聚类关系进行基因模块的划分,表达模式相似的基因将被划入同一个模块,对聚类树的分支进行剪切区分,产生不同的模块,每个颜色代表一个模块,灰色表示无法归入任何一个模块的基因。在进行初步的模块划分之后,获得初步划分的模块结果Dynamic Tree Cut,由于有些模块非常相似,我们还会再根据模块特征值的相似度对表达模式相近的模块再进行合并,获得最终划分的模块Merged dynamic。
模块特征值(module eigengene):模块中的所有基因进行PCA分析,得到的主成分1(PC1)的值,PC1相当于模块中所有基因表达量的加权,可代表该模块内基因的整体表达模式。
相似度:合并模块的参数,由模块数量决定。本次分析选用的相似度为:0.85
模块最少基因数:指每个模块至少需要包含的基因数目。本次分析选用的最少基因数为:50
下图所示,红线和蓝线为帮助模块合并的辅助线;Height指的是模块之间不相关的程度,即:不相关 = 1 - 相关性。
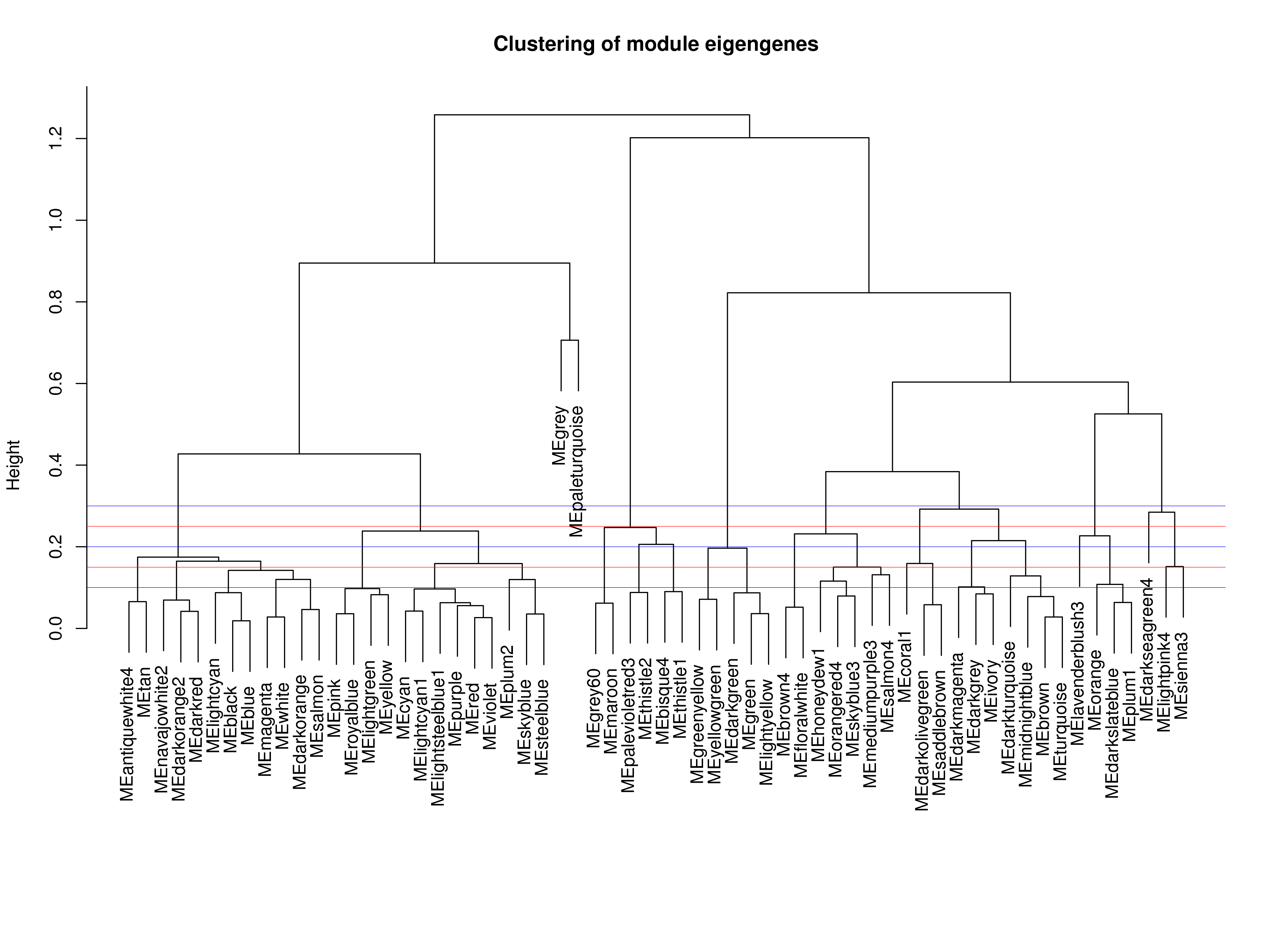 |
| Fig 4-3-1 模块特征值聚类图 |
下图所示,1)Dynamic Tree Cut为根据聚类结果划分的模块;2)Merged dynamic为根据模块相似度对表达模式相似的模块进行合并后的模块划分,之后的分析按照合并后的模块进行;3)对于树图,纵向距离代表两个节点间(基因间)的距离,横向距离无意义。
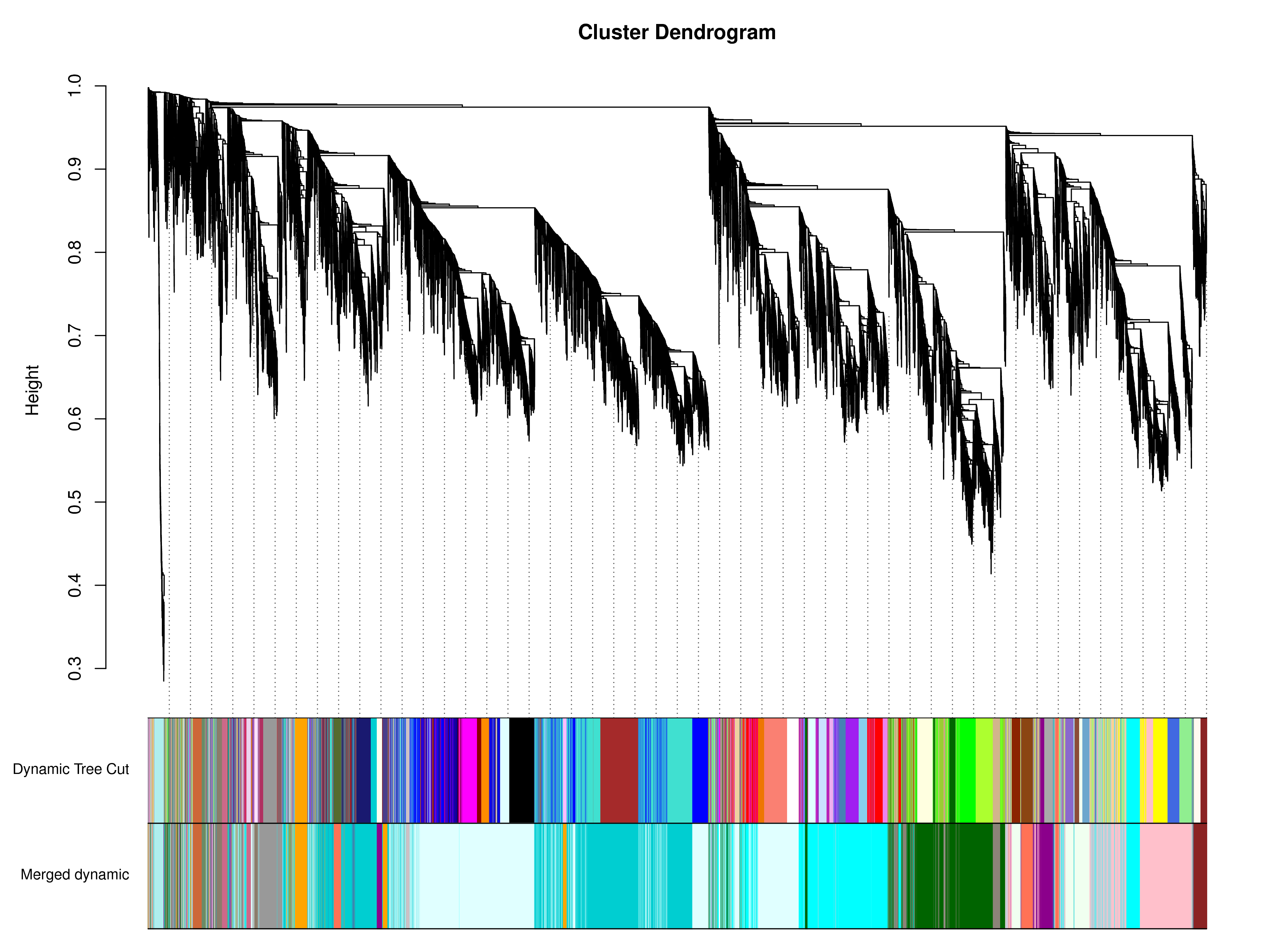 |
| Fig 4-3-2 模块层级聚类图 |
Tab 4-3-1 基因-模块对应关系列表 (仅展示前10行)
| Gene | Module | CK-1 | CK-2 | CK-3 | T36-1 | T36-2 | T36-3 | T60-1 | T60-2 | T60-3 | T72-1 | T72-2 | T72-3 |
|---|
| MSTRG.10 | pink | 1.48 | 1.10 | 1.45 | 2.86 | 1.47 | 1.69 | 1.29 | 1.44 | 2.42 | 2.41 | 2.31 | 1.43 |
| MSTRG.10026 | orange | 28.52 | 29.64 | 28.67 | 33.77 | 33.83 | 32.78 | 28.93 | 33.47 | 34.70 | 30.92 | 22.07 | 32.68 |
| MSTRG.10033 | pink | 43.46 | 43.27 | 8.30 | 11.02 | 5.75 | 5.15 | 3.68 | 6.32 | 4.48 | 0.92 | 1.38 | 2.37 |
| MSTRG.1005 | darkturquoise | 4.98 | 6.32 | 8.76 | 8.32 | 9.88 | 9.84 | 9.72 | 9.30 | 11.10 | 6.54 | 7.09 | 9.04 |
| MSTRG.10074 | darkturquoise | 3.01 | 2.87 | 3.79 | 3.79 | 3.80 | 4.59 | 5.17 | 4.41 | 5.59 | 3.33 | 2.35 | 6.03 |
| MSTRG.10085 | pink | 4.47 | 4.39 | 1.97 | 0.44 | 0.71 | 0.55 | 1.00 | 0.53 | 0.63 | 1.45 | 1.60 | 1.35 |
| MSTRG.10087 | pink | 12.25 | 12.41 | 7.77 | 1.32 | 1.63 | 1.22 | 1.22 | 1.00 | 1.46 | 4.77 | 1.34 | 3.76 |
| MSTRG.10094 | brown4 | 2.93 | 4.85 | 14.58 | 12.72 | 26.01 | 11.32 | 22.22 | 24.90 | 22.47 | 9.01 | 11.67 | 14.41 |
| MSTRG.1011 | cyan | 0.00 | 0.00 | 1.16 | 11.13 | 24.14 | 9.47 | 38.52 | 23.44 | 26.52 | 38.54 | 31.64 | 40.38 |
| MSTRG.1012 | honeydew1 | 0.21 | 1.81 | 3.36 | 3.80 | 5.85 | 2.61 | 6.59 | 4.17 | 4.10 | 2.74 | 4.48 | 4.19 |
下图中,横坐标表示每个模块,纵坐标表示基因数量。
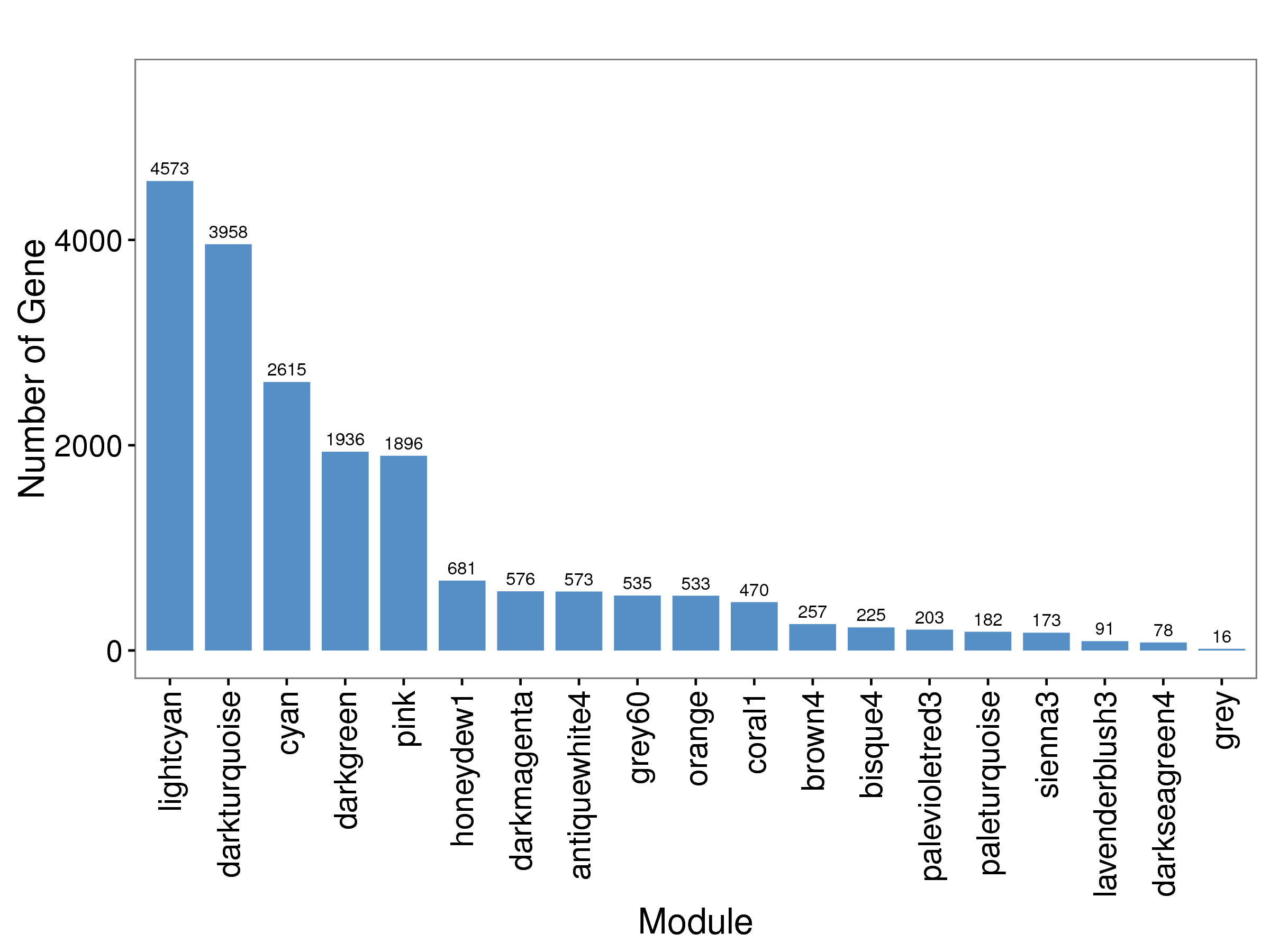 |
| Fig 4-3-3 各模块基因数柱状图 |
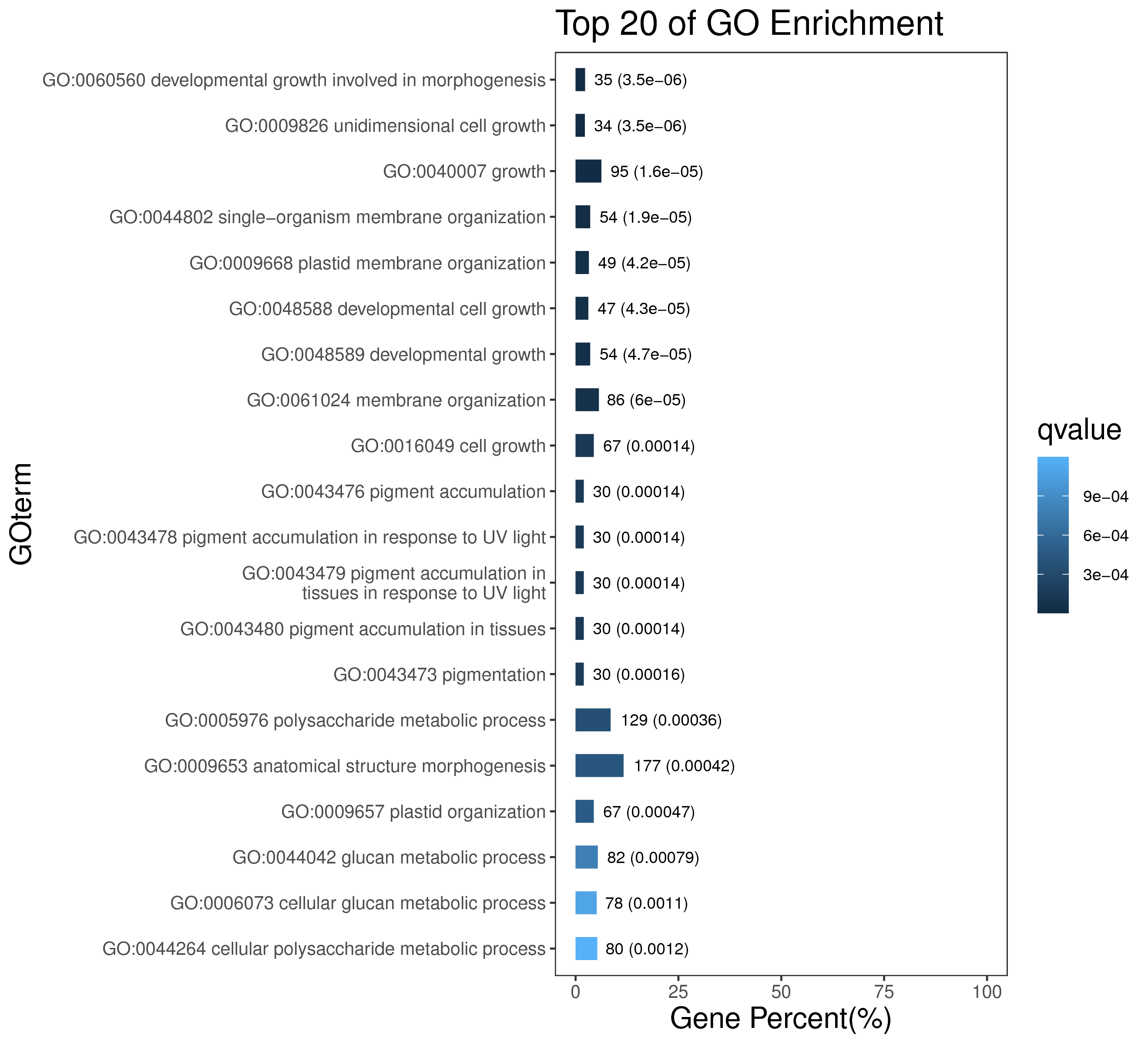
5 模块概况
每个模块具有特定的表达模式,我们对获得的模块两两之间或是样品与模块之间进行相关性和聚类分析,了解模块的具体情况。若有性状等表型信息,还可以对分析模块与性状间的关系,找出与性状最相关的模块,并进一步寻找hub基因,同时,我们也提供TF因子的信息,以便于挖掘模块内调控关系的规律。
5.1 模块间相关性分析
将所有模块两两间进行相关性分析,并绘制热图。
如下图,每一行和列代表一个模块,方块里的数字为两个模块的pearson相关性系数,括号里数字为P值。方块颜色越深(越红或越绿),相关性越强;方块颜色越浅,相关性越弱。两个模块相关性的检验值P值通过student`s T test计算,P值越小,说明两个模块间相似度越高。
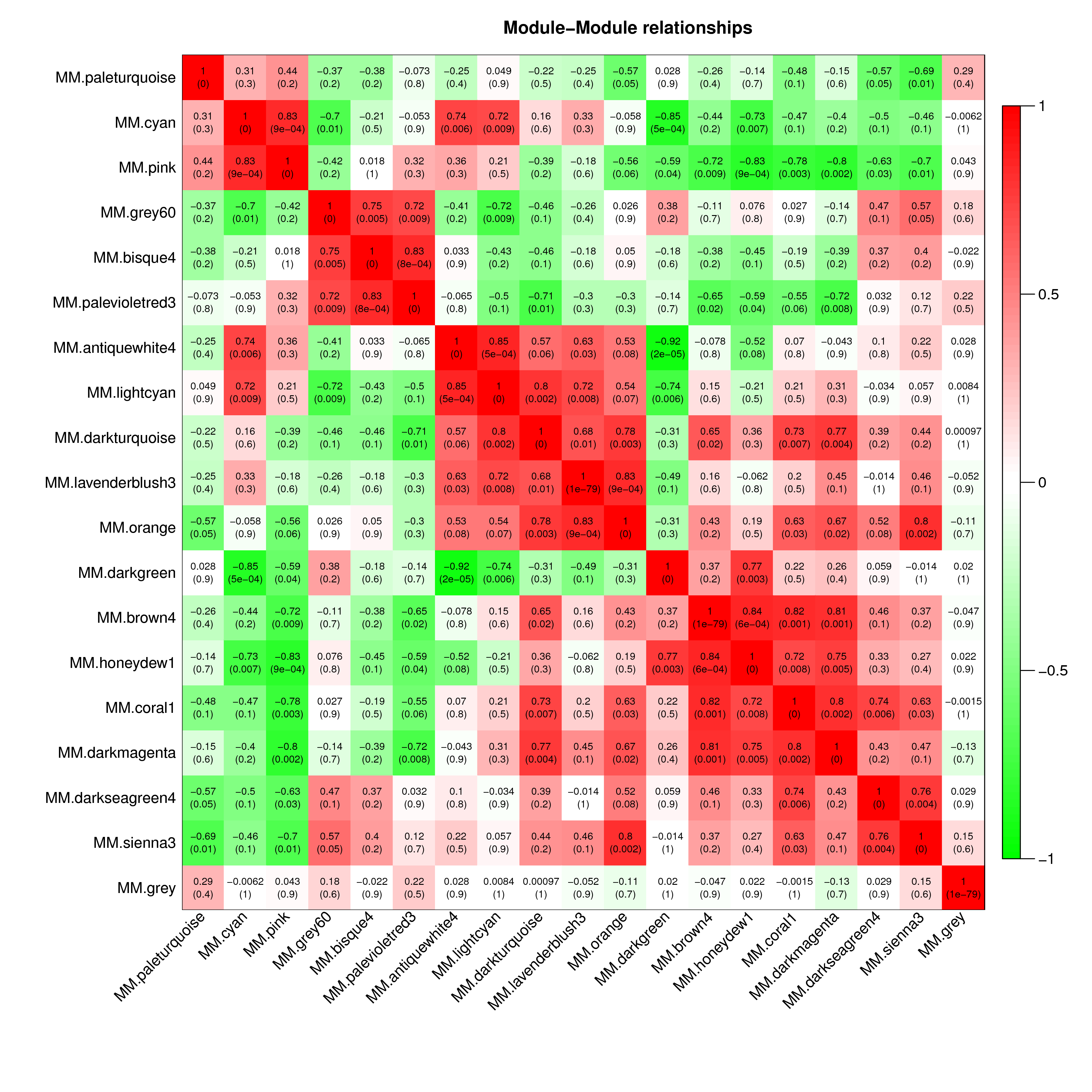 |
| Fig 5-1-1 模块间相关性热图 |
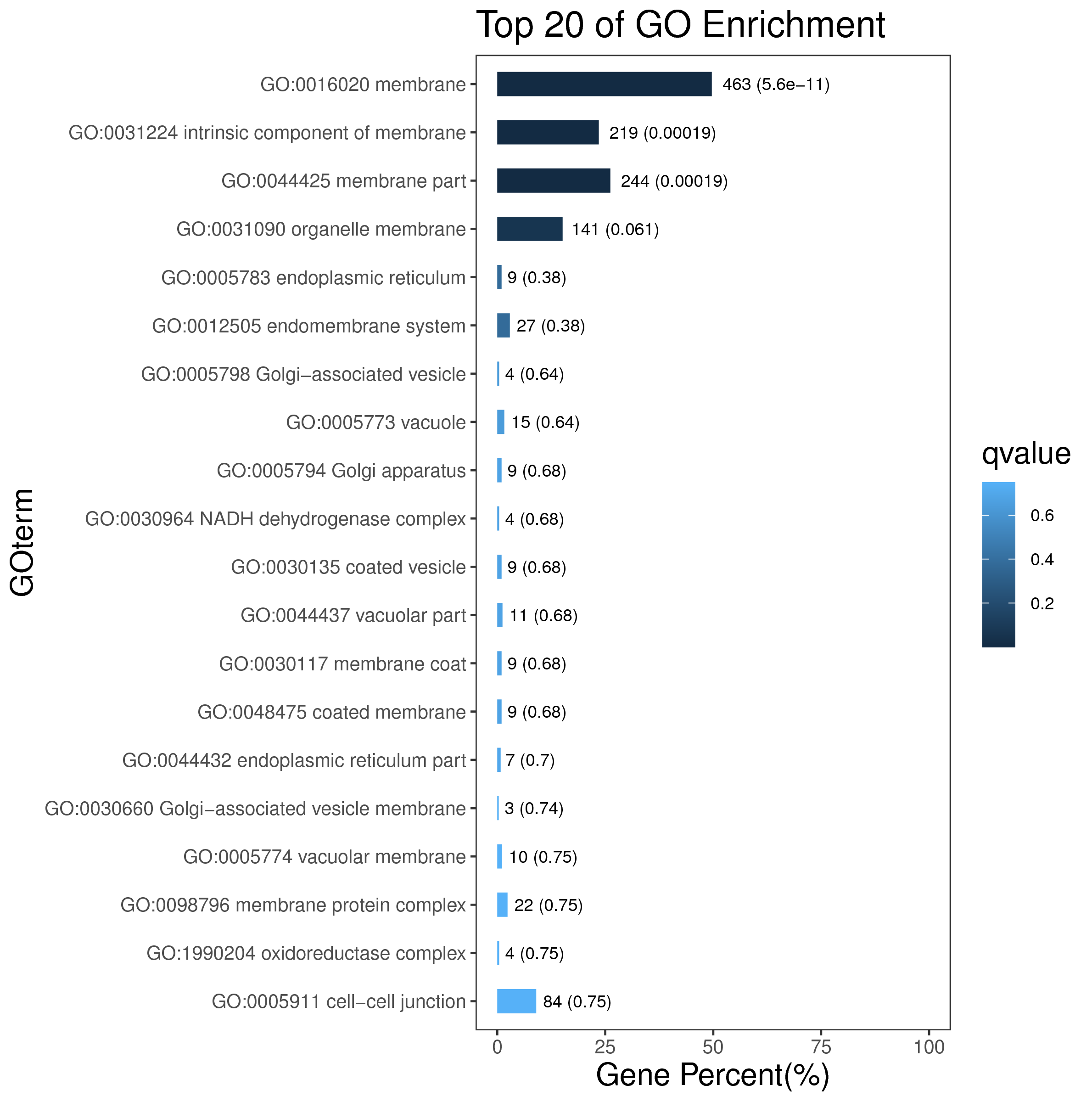
5.2 模块基因相关性分析
将模块内的基因进行聚类,并绘制热图。
如下图,每一行和列代表一个基因,每个点的颜色越深(白→黄→红)代表行和列对应的两个基因间的连通性越强,pearson相关性越强。P值采用student's t test计算所得,P值越小代表基因与模块相关性的显著性越强。
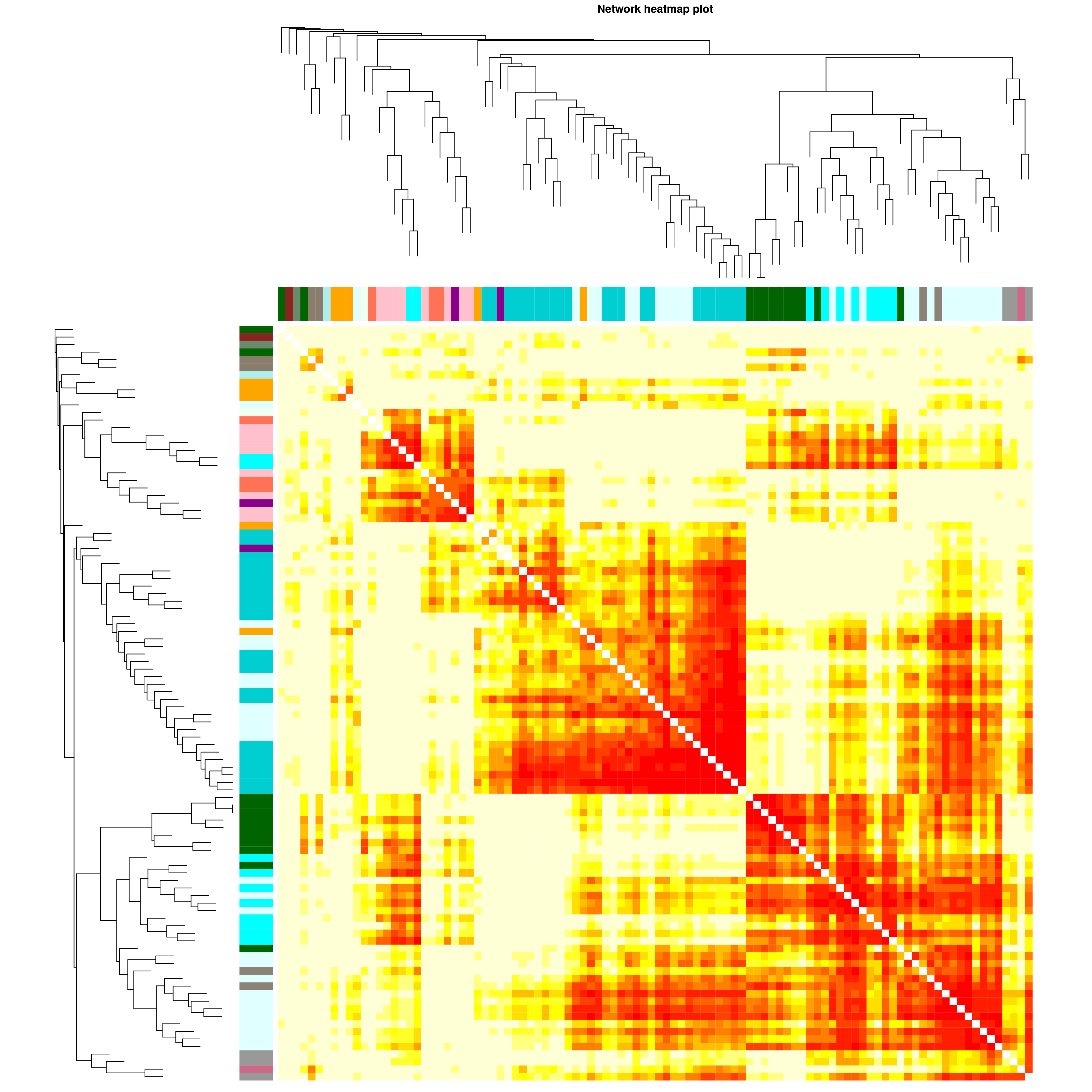 |
| Fig 5-2-1 模块基因相关性热图 |
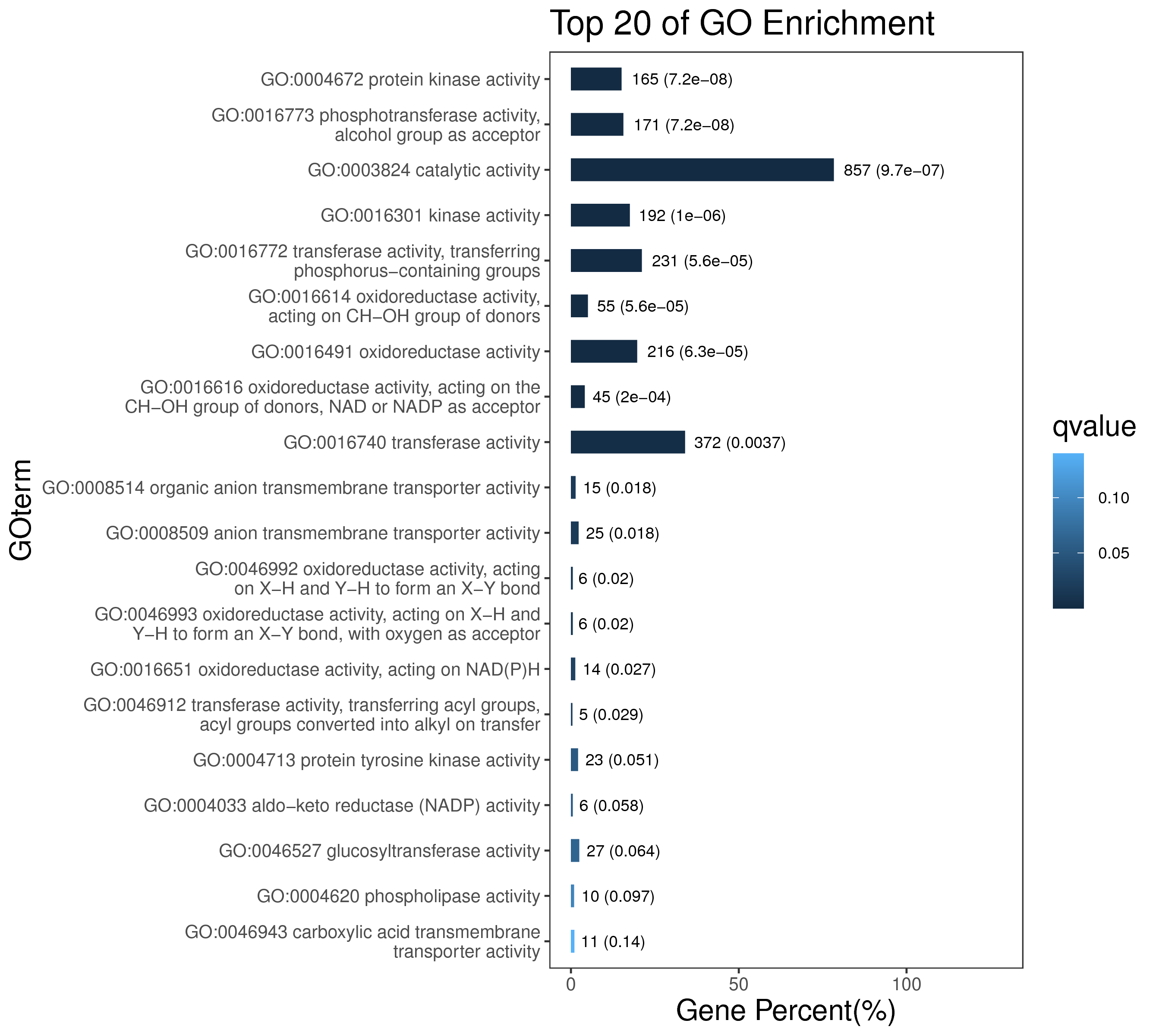
5.3 样本表达模式分析
将模块基因在各个样本中的表达模式用模块特征值来展示,并绘制样本表达模式热图。通过样本表达模式热图,我们可以找出与特定样本显著相关的模块,从而后续可选择相应的模块进行进一步的研究。模块特征值相当于模块中所有基因表达量的加权综合值。因此,模块特征值在各个样本中的数值,反映了模块中所有基因在各个样本中的综合表达水平。
如下图,横坐标为样本,纵坐标为模块,用模块特征值作图。红色代表高表达量,绿色代表低表达量。该图能直观反映各模块在各个样本中的表达模式。
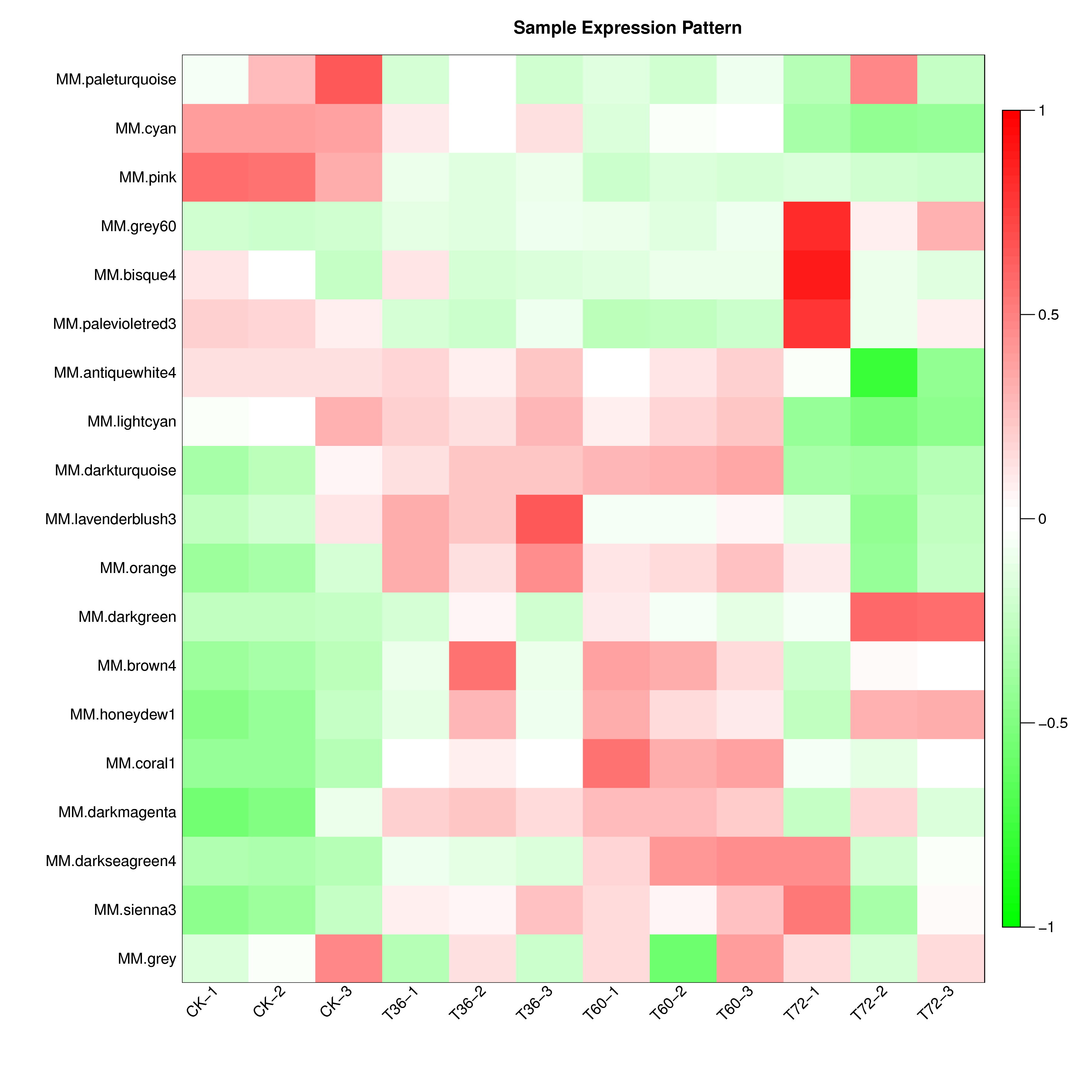 |
| Fig 5-3-1 样本表达模式热图 |
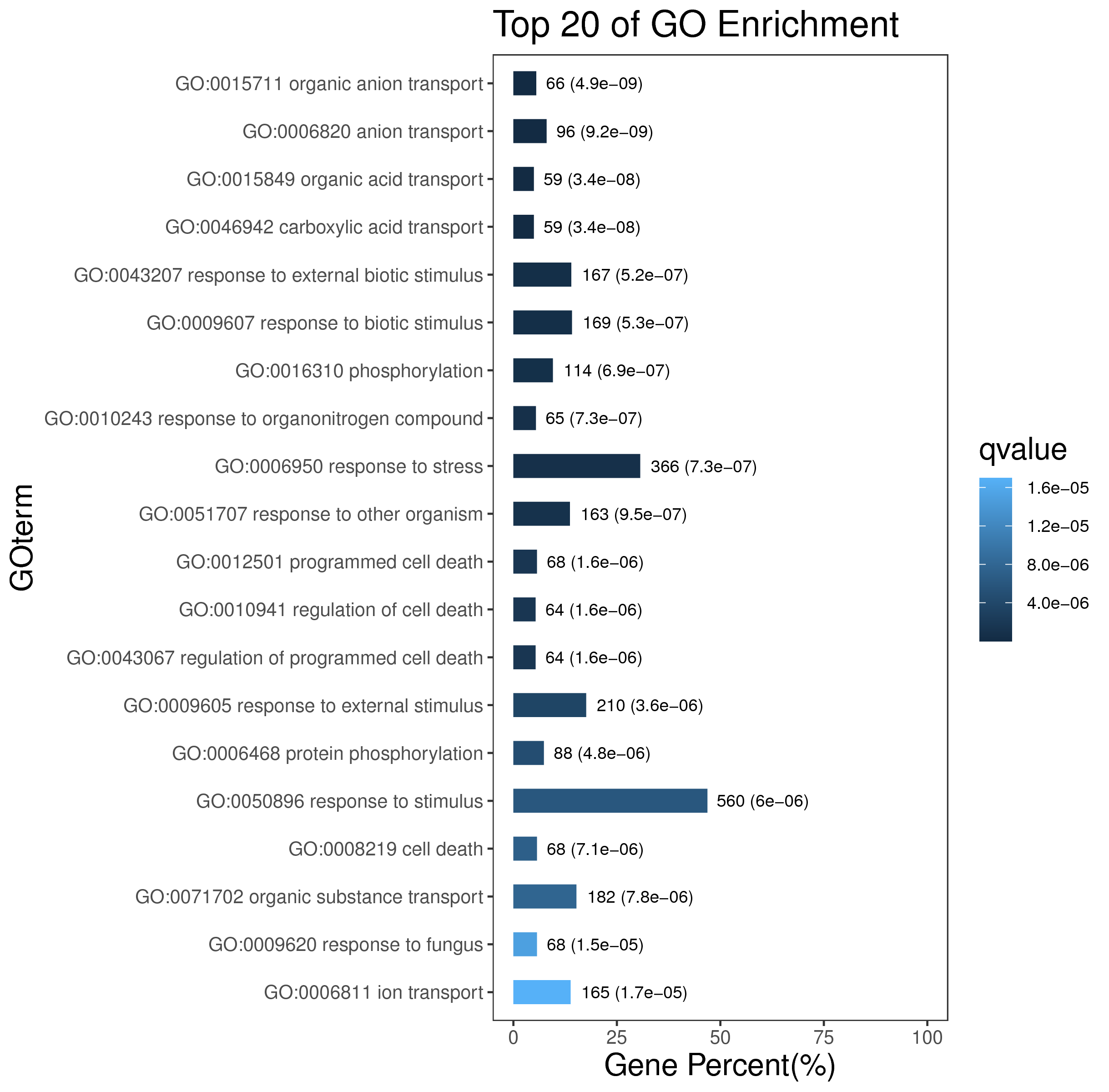
5.4 各模块hub基因挖掘
针对每个模块,我们还可以在模块中进一步挖掘hub基因,hub基因可以通过模块相关性程度MM和模块内连通性K.in(即表中All.kWithin)进行寻找,通常MM、K.in的绝对值越高,表明该基因具有重要意义。我们对所有模块的基因都计算MM和K.in值,后续可以针对该结果进一步寻找到各模块中的hub基因。
Tab 5-4-1 hub相关性结果 (仅展示前10行)
| Gene | moduleColors | All.kWithin | MM.paleturquoise | MM.cyan | MM.pink | MM.grey60 | MM.bisque4 | MM.palevioletred3 | MM.antiquewhite4 | MM.lightcyan | MM.darkturquoise | MM.lavenderblush3 | MM.orange | MM.darkgreen | MM.brown4 | MM.honeydew1 | MM.coral1 | MM.darkmagenta | MM.darkseagreen4 | MM.sienna3 | MM.grey | MM.paleturquoise.pvalue | MM.cyan.pvalue | MM.pink.pvalue | MM.grey60.pvalue | MM.bisque4.pvalue | MM.palevioletred3.pvalue | MM.antiquewhite4.pvalue | MM.lightcyan.pvalue | MM.darkturquoise.pvalue | MM.lavenderblush3.pvalue | MM.orange.pvalue | MM.darkgreen.pvalue | MM.brown4.pvalue | MM.honeydew1.pvalue | MM.coral1.pvalue | MM.darkmagenta.pvalue | MM.darkseagreen4.pvalue | MM.sienna3.pvalue | MM.grey.pvalue |
|---|
| MSTRG.10 | pink | 5.46397549215724 | -0.16719700747642 | -0.337580314122616 | -0.410754958601017 | 0.369612542739277 | 0.443285994652453 | 0.0983641816950861 | -0.137602990708348 | -0.154894679737529 | 0.00362918966792092 | 0.148708728252259 | 0.373733904839757 | 0.0629565573129345 | -0.0693842291625379 | 0.0134437844146814 | 0.123076537257278 | 0.306734100774773 | 0.345945402982086 | 0.385902317244637 | -0.0598769592785395 | 0.603494316976465 | 0.283208367635661 | 0.184709577143796 | 0.237009076487996 | 0.148921446922266 | 0.761027671926053 | 0.669772411726138 | 0.630747634091149 | 0.991068947893889 | 0.64461527780863 | 0.231412967525721 | 0.845883702637624 | 0.830341038538833 | 0.966923658168025 | 0.703150910741126 | 0.332175408265432 | 0.270682685855186 | 0.215354707945247 | 0.853348675182016 |
| MSTRG.10026 | orange | 15.0273452469243 | -0.645800976323327 | 0.131561590518916 | -0.250961048364513 | -0.024418016785026 | 0.0065053394191954 | -0.167830043643816 | 0.630057926435662 | 0.498234076241073 | 0.595254598416674 | 0.600338252167367 | 0.714450514933406 | -0.362473962999306 | 0.296097492477594 | 0.0366671372764417 | 0.392178902305716 | 0.257189824032408 | 0.431686773307903 | 0.597098391690553 | 0.0106621506539283 | 0.0233009960786901 | 0.683591535853054 | 0.431410939048934 | 0.939956532867056 | 0.983991669574431 | 0.602104090724902 | 0.0280953661927769 | 0.0992361442751188 | 0.0411572297005589 | 0.0390234005951131 | 0.00903614415641431 | 0.246890240414289 | 0.350058222968449 | 0.909926030950305 | 0.207342842960889 | 0.419673481428106 | 0.161125010169229 | 0.0403739721442489 | 0.973765090384632 |
| MSTRG.10033 | pink | 310.721790768217 | 0.180104153824894 | 0.726096468856307 | 0.907405462063653 | -0.434846224911251 | 0.050303168338137 | 0.216690750426688 | 0.334986579429324 | 0.129007092616242 | -0.389334393098407 | -0.245774819690225 | -0.509206680244224 | -0.511025161331683 | -0.587460469539468 | -0.7266972173909 | -0.644457669576175 | -0.765250443857159 | -0.541450188752566 | -0.673949866879735 | -0.182488705753039 | 0.575393382145007 | 0.00749861556529367 | 4.58087694737703e-05 | 0.157740184384116 | 0.876623759004795 | 0.498743701678154 | 0.287157946847645 | 0.689461903143947 | 0.210950920106872 | 0.44130424296626 | 0.0908606838673348 | 0.0895203143626261 | 0.044587618294843 | 0.00742502182363931 | 0.0236850196485295 | 0.00372691163186615 | 0.0690415989571355 | 0.0162515968203475 | 0.570259463364988 |
| MSTRG.1005 | darkturquoise | 261.811981147371 | -0.192361817973044 | -0.197611363472253 | -0.643451975047021 | -0.190192561337912 | -0.532252061895802 | -0.657172231584783 | 0.198441184436811 | 0.481686138554917 | 0.849192471669378 | 0.545715048500565 | 0.675698189950474 | 0.111816909155841 | 0.707754415677443 | 0.646755317745421 | 0.765513902593516 | 0.801132833884391 | 0.417036987682782 | 0.532772762792331 | 0.248497306569222 | 0.549201244762899 | 0.538137969684776 | 0.0239755205142244 | 0.553800219753862 | 0.0748523761688734 | 0.0202291569164306 | 0.536397803037756 | 0.112819475920811 | 0.000474203533309697 | 0.0664558924775746 | 0.0158730879561094 | 0.729361610446715 | 0.01002031236322 | 0.0230309328065903 | 0.00370783013905559 | 0.00173548446601121 | 0.177418850170532 | 0.0745147894713737 | 0.436097249754895 |
| MSTRG.10074 | darkturquoise | 18.8990747719072 | -0.504690467846375 | -0.262411884123078 | -0.504407336316124 | 0.0401769759903475 | -0.335926003763676 | -0.366668802699115 | 0.152431882003634 | 0.214089562251641 | 0.540724256415519 | 0.26844865705615 | 0.464307581510198 | 0.176972390144304 | 0.426907559962802 | 0.501432310823434 | 0.668208116281076 | 0.395129998161537 | 0.460924166783165 | 0.538479061937311 | 0.289581036102161 | 0.0942478915071934 | 0.409956262967005 | 0.0944630295679768 | 0.901339463959643 | 0.285723860597043 | 0.241054877998226 | 0.636255933959114 | 0.504037281864249 | 0.0694885154323461 | 0.398864838717775 | 0.128351049470203 | 0.582163753491679 | 0.166332023499653 | 0.0967435925527612 | 0.0175414292098581 | 0.20363957403102 | 0.131528784047474 | 0.0708833558995712 | 0.361261880528056 |
| MSTRG.10085 | pink | 154.218264242274 | 0.360971093001729 | 0.518710730759613 | 0.896531929363748 | -0.179840161456414 | 0.145597713259158 | 0.450456796253957 | 0.0290847950126714 | -0.187311551016691 | -0.671434250208853 | -0.521898291693249 | -0.779290277378527 | -0.268902872719401 | -0.686667405801078 | -0.676590412797026 | -0.758837142181421 | -0.90570357894288 | -0.565339061437244 | -0.725621891457708 | 0.0328079062352891 | 0.249000882127243 | 0.0840029817360041 | 7.83211965504143e-05 | 0.575962881689061 | 0.651629365904816 | 0.141682137984007 | 0.928504806279814 | 0.559932528174219 | 0.0168078107026441 | 0.0817840372935295 | 0.00280896708397081 | 0.398036505436902 | 0.013644451766092 | 0.0156824424478911 | 0.00421482305795469 | 5.00288739287409e-05 | 0.0554172598416934 | 0.00755713825578735 | 0.919377552089288 |
| MSTRG.10087 | pink | 182.135114120653 | 0.34517813863206 | 0.626784846689794 | 0.943326283024287 | -0.149105432836483 | 0.191290099385311 | 0.53530286469974 | 0.19713315608285 | -0.0559289699240517 | -0.599216127367693 | -0.380636401708537 | -0.679650612252391 | -0.421990289118142 | -0.754754473138781 | -0.789175782845866 | -0.798071607400294 | -0.942391214003754 | -0.534265860099897 | -0.62384218633893 | 0.151302725412966 | 0.271818048988885 | 0.0291751446871998 | 4.18493600945728e-06 | 0.643722763234015 | 0.551471382262117 | 0.0728892880167713 | 0.539141869265351 | 0.86293473697344 | 0.0394874751629767 | 0.222218810441154 | 0.0150413415036728 | 0.171799023571335 | 0.00454967583636515 | 0.00227420454707458 | 0.00186292570865751 | 4.53452825374845e-06 | 0.073552554808143 | 0.030171162918531 | 0.638787094099933 |
| MSTRG.10094 | brown4 | 48.6980122455299 | -0.166015783849706 | -0.298894833987972 | -0.653937038058816 | -0.169030149374153 | -0.430069569859702 | -0.653509856377204 | 0.0903370499278559 | 0.347120643349756 | 0.783180633848058 | 0.247976463086513 | 0.501047260497091 | 0.203775331047914 | 0.927311319278625 | 0.746183882792247 | 0.829804420193713 | 0.816596976589981 | 0.53747132087627 | 0.417684425552032 | 0.115857317203746 | 0.606091667731471 | 0.345306351483752 | 0.0210710668955994 | 0.599471848158582 | 0.162875247811975 | 0.0211841134275396 | 0.780087474725695 | 0.268948922340308 | 0.00258820206150755 | 0.437091097584232 | 0.097041434308027 | 0.525269148398159 | 1.41328705003176e-05 | 0.00531772663286092 | 0.000838787697824678 | 0.001190419613648 | 0.0715156342924433 | 0.17667784824343 | 0.719923945087724 |
| MSTRG.1011 | cyan | 248.330335851192 | -0.414840200751593 | -0.943551273927608 | -0.835698987941611 | 0.650056242424747 | 0.193642598745467 | 0.00112913184395867 | -0.553509801030212 | -0.573332180617222 | 0.0165900468858819 | -0.299269227019724 | 0.155899898794894 | 0.734536588709434 | 0.580286182624015 | 0.757787891848333 | 0.651501235731118 | 0.432849554166202 | 0.648441550815165 | 0.573624166170713 | 0.149270611609682 | 0.179947568470958 | 4.10409582552642e-06 | 0.00071066674144055 | 0.0221143562501068 | 0.546493363798171 | 0.997221281826354 | 0.0619043248054487 | 0.0513127461922039 | 0.959187910207691 | 0.344673002015155 | 0.628504287216621 | 0.00651347506050197 | 0.0479194810519425 | 0.00429903686019003 | 0.0217215970399549 | 0.159873976789685 | 0.0225593289483584 | 0.051166975600108 | 0.643351267730829 |
| MSTRG.1012 | honeydew1 | 36.0257712857298 | -0.0499803544167186 | -0.549219140676997 | -0.757663026706578 | 0.00475773068891355 | -0.366277973642613 | -0.577904760825604 | -0.256904530681579 | 0.0251266957097712 | 0.533199574306314 | 0.0950366991097154 | 0.338649499359058 | 0.505505745178709 | 0.8734727813477 | 0.877174381731288 | 0.771887318937638 | 0.805207117146991 | 0.358145875214185 | 0.364657511822361 | 0.186407186776756 | 0.877410224582107 | 0.0643820945626967 | 0.00430914280971609 | 0.988291875503047 | 0.241595074279126 | 0.0490631685083435 | 0.420207607138773 | 0.938216795672042 | 0.074238853270783 | 0.768914350013554 | 0.281589327812119 | 0.0936302424438273 | 0.000205739035913186 | 0.000178506346088667 | 0.00326839143829153 | 0.00157636221262207 | 0.252997216844726 | 0.243842472092172 | 0.5618631782611 |
5.5 各模块TF因子分析
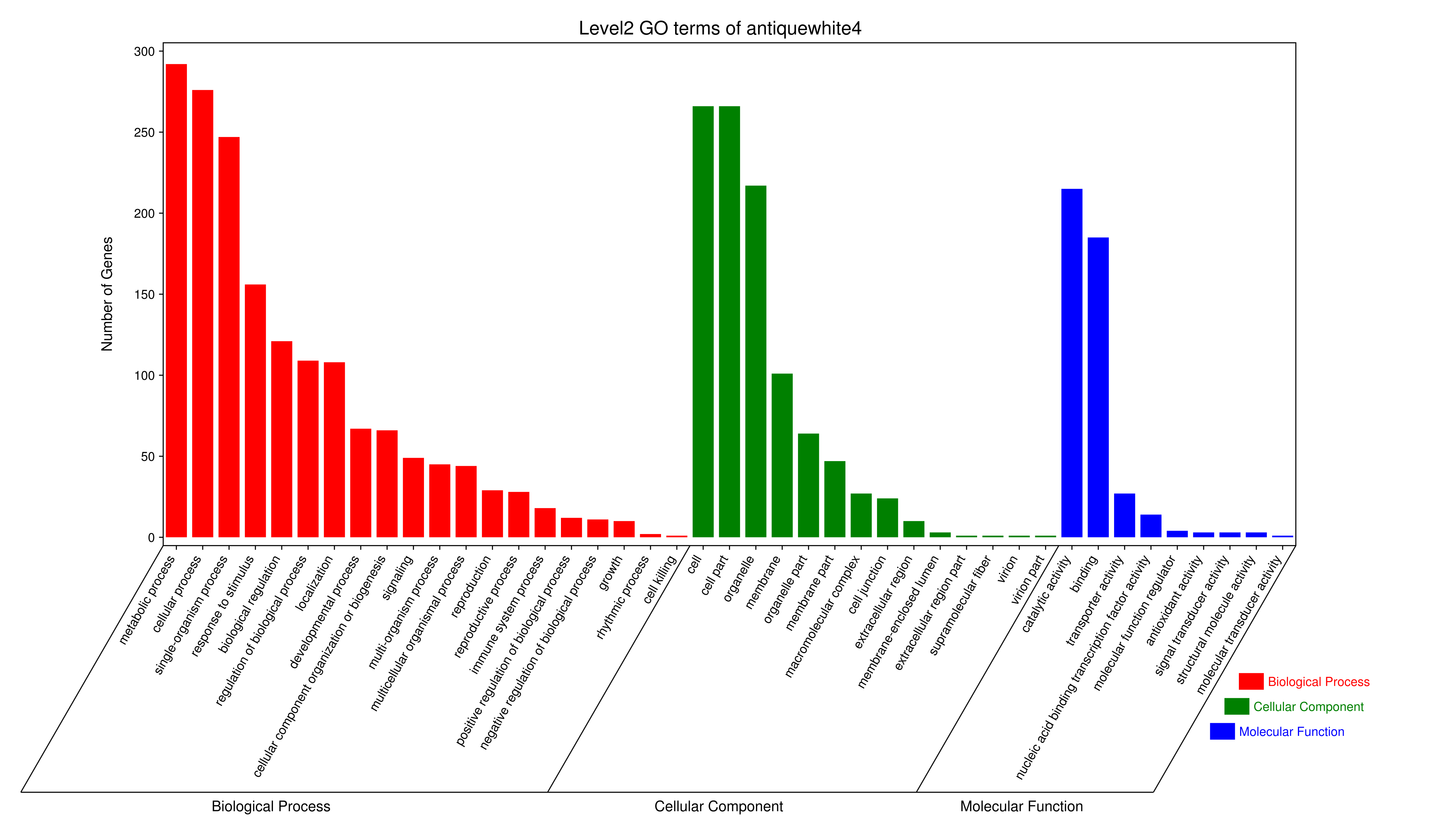
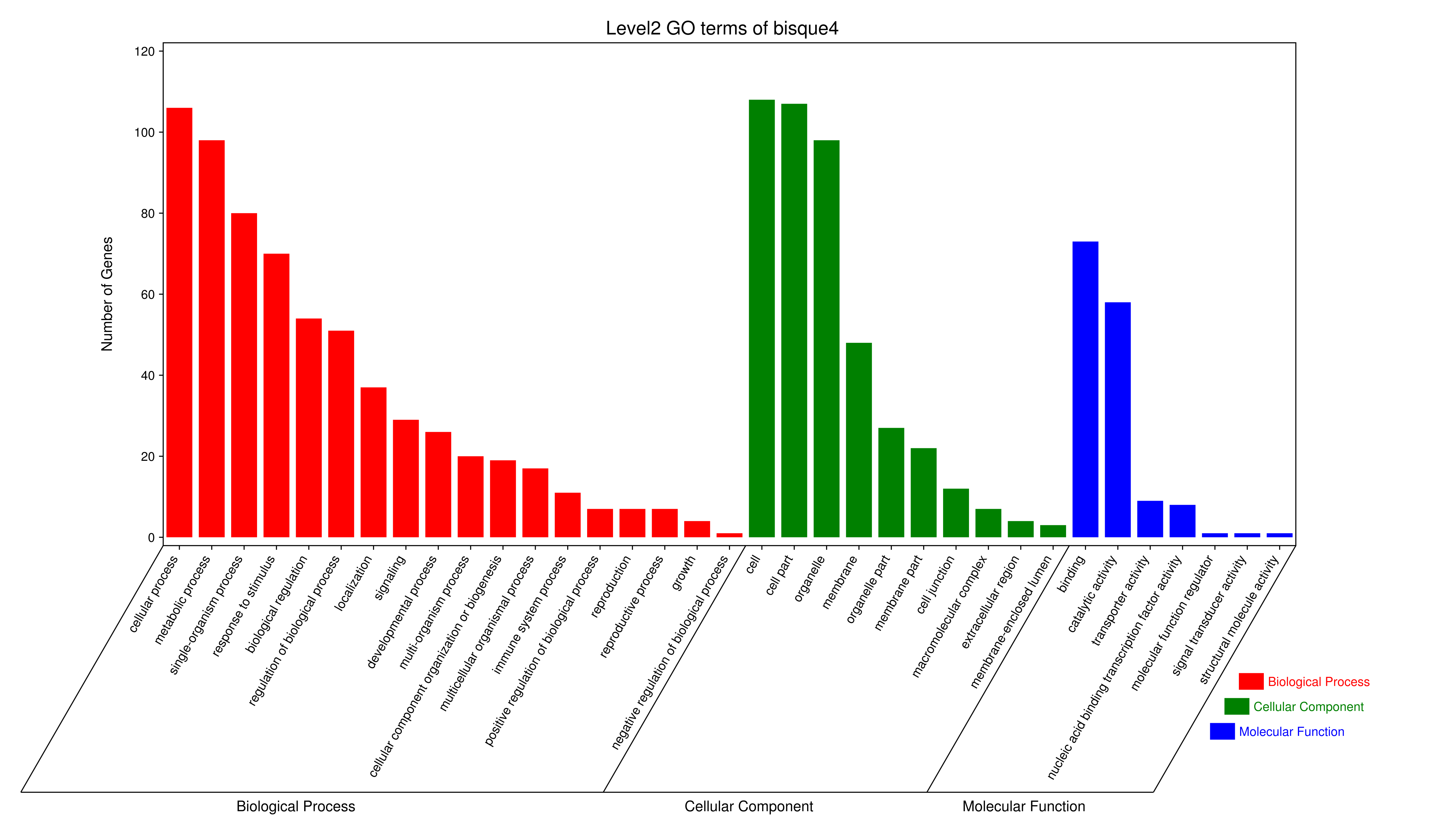
转录因子(TF)对于转录调控起到重要作用,我们寻找各模块中的转录因子,若分析的物种是植物或者动物,我们将预测的蛋白序列同相应的TF数据库(plant TFdb[3]/animal TFdb[4])进行 hmmscan 比对,获得该物种的TF因子的信息,便于在后续构建共表达网络图时找到关键的TF因子,以便于进一步了解TF因子调控的机制。
Tab 5-5-1 TF预测结果 (仅展示前10行)
| Gene | TF family | Module |
|---|
| ncbi_112163987 | GRAS | honeydew1 |
| ncbi_112164284 | AP2-EREBP | cyan |
| ncbi_112164486 | WRKY | darkgreen |
| ncbi_112164521 | NAC | coral1 |
| ncbi_112164550 | WRKY | darkturquoise |
| ncbi_112164583 | WRKY | lightcyan |
| ncbi_112164600 | ARR-B | darkturquoise |
| ncbi_112164616 | bZIP | pink |
| ncbi_112164651 | TCP | lightcyan |
| ncbi_112164653 | ARR-B | cyan |
Tab 5-5-2 TF因子统计表
| Module | TF number |
|---|
| lightcyan | 178 |
| cyan | 167 |
| darkturquoise | 122 |
| pink | 90 |
| darkgreen | 71 |
| honeydew1 | 39 |
| antiquewhite4 | 30 |
| coral1 | 24 |
| orange | 21 |
| grey60 | 20 |
| bisque4 | 18 |
| darkmagenta | 17 |
| sienna3 | 13 |
| palevioletred3 | 12 |
| brown4 | 10 |
| darkseagreen4 | 5 |
| paleturquoise | 3 |
| lavenderblush3 | 3 |
下图中,横坐标表示每个模块,纵坐标表示转录因子的数量。
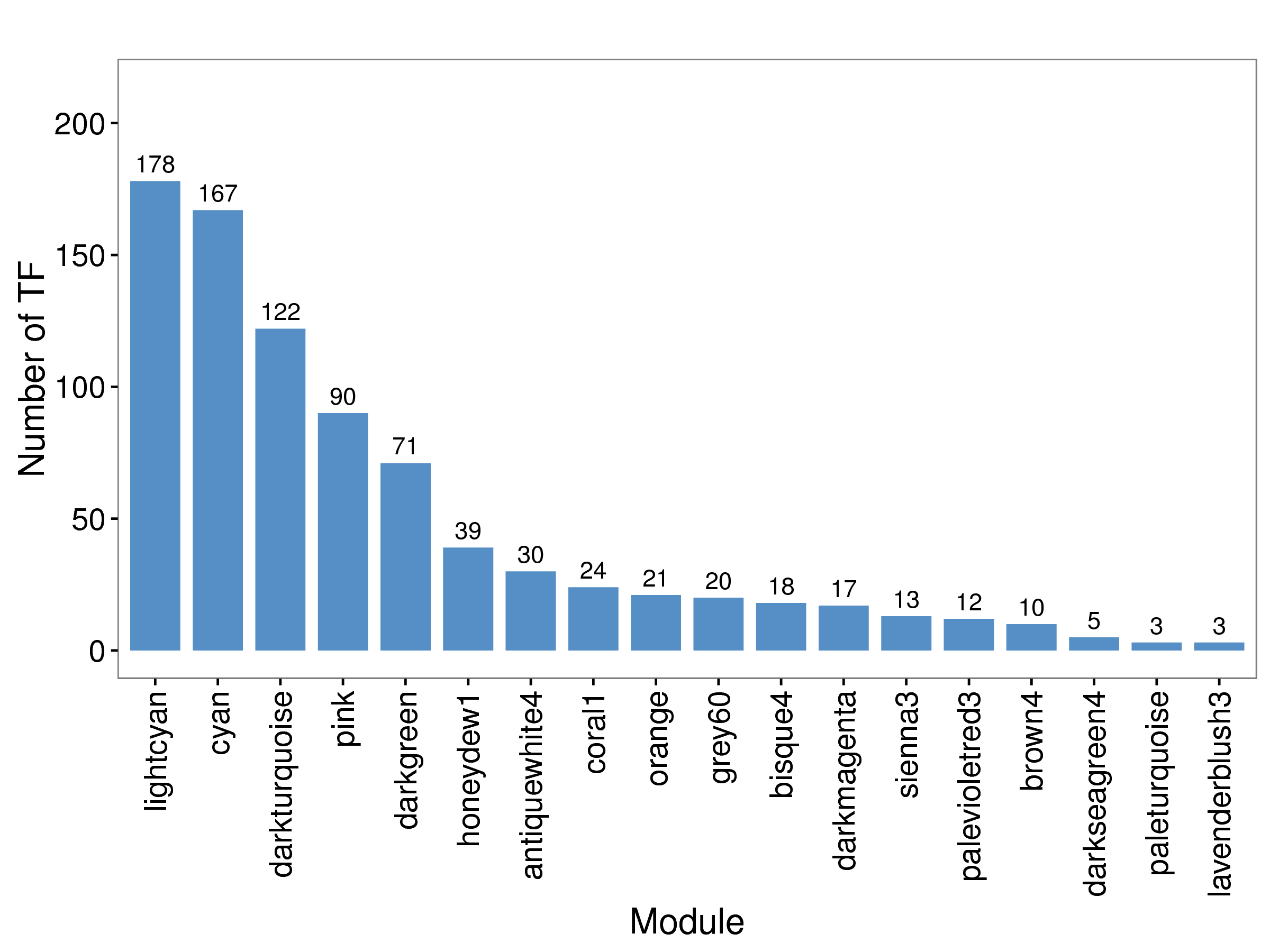 |
| Fig 5-5-1 各模块TF因子的柱状图 |
6 表达模式
6.1 模块基因总表
Tab 6-1-1 模块基因总表 (仅展示前10行)
| GeneID | Module | All.kTotal | All.kWithin | MM.paleturquoise | MM.cyan | MM.pink | MM.grey60 | MM.bisque4 | MM.palevioletred3 | MM.antiquewhite4 | MM.lightcyan | MM.darkturquoise | MM.lavenderblush3 | MM.orange | MM.darkgreen | MM.brown4 | MM.honeydew1 | MM.coral1 | MM.darkmagenta | MM.darkseagreen4 | MM.sienna3 | MM.grey | MM.paleturquoise.pvalue | MM.cyan.pvalue | MM.pink.pvalue | MM.grey60.pvalue | MM.bisque4.pvalue | MM.palevioletred3.pvalue | MM.antiquewhite4.pvalue | MM.lightcyan.pvalue | MM.darkturquoise.pvalue | MM.lavenderblush3.pvalue | MM.orange.pvalue | MM.darkgreen.pvalue | MM.brown4.pvalue | MM.honeydew1.pvalue | MM.coral1.pvalue | MM.darkmagenta.pvalue | MM.darkseagreen4.pvalue | MM.sienna3.pvalue | MM.grey.pvalue | CK-1 | CK-2 | CK-3 | T36-1 | T36-2 | T36-3 | T60-1 | T60-2 | T60-3 | T72-1 | T72-2 | T72-3 | Symbol | Description | KEGG_A_class | KEGG_B_class | Pathway | K_ID | GO Component | GO Function | GO Process | TF_family |
|---|
| MSTRG.10786 | antiquewhite4 | 2058.76098088245 | 234.332729582144 | 0.192345033091137 | -0.789847074877546 | -0.470799745089072 | 0.373754253919778 | -0.0731085988142983 | -0.056268194902974 | -0.976706028892179 | -0.787296318218886 | -0.435300104723269 | -0.589069378651897 | -0.413860240199477 | 0.930842883979297 | 0.172994047150331 | 0.605343002010505 | 0.0933332578148155 | 0.184002132986911 | 0.0191571607700432 | -0.139934342498874 | -0.0555795464693274 | 0.549236768963246 | 0.00224094888425415 | 0.122394775391805 | 0.231385534350503 | 0.821360322890045 | 0.86211038480644 | 5.1943629264324e-08 | 0.00236932067880131 | 0.157257674227702 | 0.0438635215640025 | 0.181082811921952 | 1.10842870019211e-05 | 0.590809025804316 | 0.0370007661168008 | 0.772959628467899 | 0.567010629524881 | 0.952878486260706 | 0.664464434978654 | 0.863784003734782 | 2.89 | 4.02 | 3.05 | 4.89 | 4.87 | 3.07 | 9.23 | 6.10 | 6.34 | 8.04 | 26.72 | 17.03 | -- | PREDICTED: muscle M-line assembly protein unc-89-like [Pyrus x bretschneideri] | - | - | - | - | GO:0043231//intracellular membrane-bounded organelle | - | - | - |
| MSTRG.11431 | antiquewhite4 | 54.6450296004314 | 8.40825285571775 | -0.48283639682843 | -0.0482615400924883 | -0.0970649346264394 | 0.346710631586429 | 0.345929674872081 | 0.307706732839491 | 0.40466207135888 | 0.0699800442774538 | 0.171691678198712 | 0.0979540159358844 | 0.27944367203074 | -0.205522416423414 | 0.192146750458971 | -0.0219487062737134 | 0.287103200619694 | -0.126436463962761 | 0.418417753818641 | 0.557689896824169 | 0.520070837136929 | 0.111837642480424 | 0.881599437951703 | 0.764104658448094 | 0.269553059593605 | 0.270705932144732 | 0.330565412550846 | 0.191955903614034 | 0.828903046559041 | 0.593649918596687 | 0.761998720002863 | 0.379060228561402 | 0.521645951886478 | 0.549656494405682 | 0.946020285552057 | 0.365570939957836 | 0.695385248309431 | 0.175840885773788 | 0.0595547930264455 | 0.0830512375414281 | 2.85 | 3.32 | 2.94 | 2.86 | 3.77 | 2.69 | 3.77 | 2.77 | 3.50 | 4.06 | 1.56 | 3.55 | MPT2 | mitochondrial phosphate carrier protein 2, mitochondrial [Rosa chinensis] | - | - | - | - | GO:0009536//plastid;GO:0019866//organelle inner membrane;GO:0030312//external encapsulating structure;GO:0031224//intrinsic component of membrane | - | GO:0006006//glucose metabolic process;GO:0006739//NADP metabolic process;GO:0006970//response to osmotic stress;GO:0009725//response to hormone;GO:0051234//establishment of localization | - |
| MSTRG.1162 | antiquewhite4 | 1706.03916799444 | 118.587824639664 | 0.179932787607858 | -0.679308198650942 | -0.255368528807125 | 0.547691179641409 | 0.0384301753292093 | 0.273827251303733 | -0.839157147927077 | -0.817188465938962 | -0.575345161015403 | -0.69522865780465 | -0.572400770661289 | 0.822946268423824 | -0.0236718832629048 | 0.415860139405769 | -0.0531341540958046 | -0.17629681488663 | 0.0260540306027714 | -0.0864831292124016 | 0.348674863411276 | 0.57576303774542 | 0.0151120992858209 | 0.423089066028177 | 0.0652807882624394 | 0.905611725941013 | 0.389112268476607 | 0.000642895544910312 | 0.00117258807716284 | 0.050313686861409 | 0.0120727290274685 | 0.0517796892050812 | 0.00100954107633404 | 0.941788477734983 | 0.178770738832819 | 0.869731140203655 | 0.583628327001538 | 0.93594065515924 | 0.789278618727384 | 0.266665934444098 | 0.45 | 0.75 | 0.89 | 0.18 | 0.50 | 0.15 | 0.93 | 0.46 | 0.65 | 1.13 | 1.57 | 2.02 | Os04g0119400 | pyruvate dehydrogenase E1 component subunit alpha-3, chloroplastic [Rosa chinensis] | Metabolism;Metabolism;Metabolism;Metabolism;Metabolism;Metabolism | Global and overview maps;Global and overview maps;Global and overview maps;Carbohydrate metabolism;Carbohydrate metabolism;Carbohydrate metabolism | ko01100//Metabolic pathways;ko01110//Biosynthesis of secondary metabolites;ko01200//Carbon metabolism;ko00010//Glycolysis / Gluconeogenesis;ko00620//Pyruvate metabolism;ko00020//Citrate cycle (TCA cycle) | K00161;K00161;K00161;K00161;K00161;K00161 | GO:0009526//plastid envelope;GO:0009532//plastid stroma | GO:0004738//pyruvate dehydrogenase activity | GO:0006090//pyruvate metabolic process | - |
| MSTRG.12109 | antiquewhite4 | 101.463662937402 | 12.4997149849126 | -0.349081289124499 | 0.169869684499217 | -0.0377393974663821 | 0.236554977385988 | 0.253771838121485 | 0.292414417967466 | 0.5181340478182 | 0.311566365240801 | 0.249349090844733 | 0.516526336350605 | 0.533962605355671 | -0.453173350257988 | -0.183008151086383 | -0.310435944548392 | 0.0442351336205528 | -0.055859633344309 | 0.346928028552946 | 0.572787851567176 | 0.175553457473046 | 0.266070790779971 | 0.5976330706523 | 0.907301845417477 | 0.459157745889438 | 0.426094644234734 | 0.356367502053652 | 0.0844087455505091 | 0.324218537168603 | 0.434474279883069 | 0.0855469760944896 | 0.073747295533166 | 0.139000048805271 | 0.569143543580771 | 0.326070477042335 | 0.891423616198666 | 0.863103247586444 | 0.269232635826741 | 0.0515852739274237 | 0.58524150971334 | 2.98 | 3.83 | 3.42 | 3.24 | 3.32 | 4.57 | 2.78 | 3.51 | 4.36 | 4.19 | 2.48 | 3.45 | -- | zinc finger MYM-type protein 1-like [Rosa chinensis] | - | - | - | - | GO:0043231//intracellular membrane-bounded organelle | GO:0005515//protein binding | - | - |
| MSTRG.12245 | antiquewhite4 | 199.376564531357 | 24.8978312639893 | -0.216903473968014 | 0.217451434199096 | -0.0701110583185955 | 0.0316820698989083 | 0.290259692822612 | 0.0542401141517599 | 0.628988175474489 | 0.478603196158217 | 0.476090348783 | 0.455710990804761 | 0.554974921069137 | -0.526159915099508 | 0.151056924669248 | -0.168629141317315 | 0.306883535168903 | 0.190558703745578 | 0.321541809885376 | 0.515284973450122 | 0.4618284065714 | 0.49831189839962 | 0.497200375306443 | 0.828586910385127 | 0.922136658887591 | 0.360086359146792 | 0.867040686401968 | 0.0284450462585033 | 0.115479062257905 | 0.117677185028087 | 0.136524449496537 | 0.0610736239054029 | 0.0788801824914958 | 0.639338560709717 | 0.600350908555619 | 0.331927773569635 | 0.553022861888645 | 0.30812694712026 | 0.0864329245058649 | 0.130674554886716 | 1.06 | 1.03 | 1.49 | 1.71 | 1.57 | 1.05 | 1.68 | 0.87 | 1.60 | 1.65 | 0.46 | 0.73 | rpsE | probable 37S ribosomal protein S5, mitochondrial [Rosa chinensis] | Genetic Information Processing | Translation | ko03010//Ribosome | K02988 | - | - | - | - |
| MSTRG.12586 | antiquewhite4 | 534.837665058289 | 82.8079363292945 | 0.532157647367151 | -0.23532589884363 | 0.10631366105142 | -0.096605966602373 | -0.438685545991436 | -0.121888036420232 | -0.781420143875696 | -0.534778271512551 | -0.535521475598063 | -0.589018477583406 | -0.760161139476563 | 0.680827404714321 | -0.104234827612871 | 0.28796564840339 | -0.364278393315044 | -0.194457035574626 | -0.508725974001952 | -0.679693939692834 | -0.0630867045964235 | 0.0749137000444957 | 0.461563100432524 | 0.742271648792881 | 0.76519237673457 | 0.153688310237396 | 0.705904134063995 | 0.00268639904508733 | 0.0732243020403194 | 0.0727499904156796 | 0.0438863015106805 | 0.00411034748719766 | 0.0148000243484473 | 0.747164473363101 | 0.364068060046122 | 0.244370040460089 | 0.544774345535625 | 0.0912172529550893 | 0.0150324055697609 | 0.845568476401307 | 2.14 | 2.46 | 2.18 | 1.43 | 1.92 | 1.68 | 1.61 | 1.92 | 1.61 | 1.16 | 3.50 | 3.05 | -- | hypothetical protein RchiOBHm_Chr3g0490531 [Rosa chinensis] | - | - | - | - | - | - | - | - |
| MSTRG.12762 | antiquewhite4 | 1222.72257370144 | 136.197126289498 | -0.225958103538644 | 0.737781160245119 | 0.444387769032224 | -0.383387782240677 | 0.0841540763842799 | 0.0228623206463631 | 0.880634625253432 | 0.680821716398784 | 0.349037784551891 | 0.55822325327497 | 0.376895267555669 | -0.835887858690047 | -0.0984493316622386 | -0.55325013030362 | -0.172182861866582 | -0.183847440626352 | -0.0904395427252867 | 0.0662107342203426 | -0.0612395418570098 | 0.480088599169307 | 0.00616190972343925 | 0.147794055766517 | 0.218616189591323 | 0.79484518701996 | 0.943776449651863 | 0.000155677414452104 | 0.0148011839042809 | 0.266134459340435 | 0.0592595365905772 | 0.227174307914541 | 0.000706828582687819 | 0.760826123020425 | 0.0620523675751168 | 0.592577868973437 | 0.56734236097641 | 0.779843388757628 | 0.838008182661554 | 0.850044365931033 | 45.03 | 44.40 | 40.30 | 43.42 | 46.66 | 43.74 | 36.50 | 41.26 | 42.45 | 40.02 | 29.73 | 34.37 | MLO2 | MLO-like protein 3 [Rosa chinensis] | - | - | - | - | GO:0005911//cell-cell junction;GO:0016020//membrane;GO:0043231//intracellular membrane-bounded organelle | - | GO:0002252//immune effector process;GO:0006605//protein targeting;GO:0006812//cation transport;GO:0009404//toxin metabolic process;GO:0009617//response to bacterium;GO:0009696//salicylic acid metabolic process;GO:0009755//hormone-mediated signaling pathway;GO:0009863//salicylic acid mediated signaling pathway;GO:0010053//root epidermal cell differentiation;GO:0010243//response to organonitrogen compound;GO:0010260//organ senescence;GO:0015833//peptide transport;GO:0031347//regulation of defense response;GO:0035556//intracellular signal transduction;GO:0042743//hydrogen peroxide metabolic process;GO:0043067//regulation of programmed cell death;GO:0046942//carboxylic acid transport;GO:0048468//cell development;GO:0050832//defense response to fungus | - |
| MSTRG.1286 | antiquewhite4 | 2894.15645203472 | 37.5015724566461 | 0.0307150642451634 | 0.860799453898154 | 0.503201140974355 | -0.671421618288681 | -0.227492604915585 | -0.248878187543506 | 0.923845923539288 | 0.910053904750661 | 0.565956968007369 | 0.477506219187179 | 0.308310657005444 | -0.887543820099833 | -0.0732315754851193 | -0.486138884814125 | 0.00907806939420662 | -0.0492413984809363 | -0.0479206802563757 | -0.0876165537469658 | -0.0101312988565171 | 0.924507146859949 | 0.000324297005915343 | 0.095383258627676 | 0.0168106384664688 | 0.477030918886292 | 0.435371165302454 | 1.7732950070359e-05 | 3.98032992463316e-05 | 0.0550920472777314 | 0.116435287591881 | 0.329567877775517 | 0.00011694482114052 | 0.821064114253407 | 0.109050197934977 | 0.977661893239046 | 0.879210911326767 | 0.882430539475406 | 0.786572922000403 | 0.975070918606831 | 19.90 | 20.48 | 21.17 | 18.31 | 17.37 | 18.76 | 17.33 | 20.51 | 20.06 | 12.13 | 6.91 | 9.40 | PFP-BETA | probable LRR receptor-like serine/threonine-protein kinase At1g53430 isoform X5 [Rosa chinensis] | Metabolism;Metabolism;Metabolism;Metabolism;Metabolism | Global and overview maps;Global and overview maps;Carbohydrate metabolism;Carbohydrate metabolism;Carbohydrate metabolism | ko01100//Metabolic pathways;ko01110//Biosynthesis of secondary metabolites;ko00010//Glycolysis / Gluconeogenesis;ko00051//Fructose and mannose metabolism;ko00030//Pentose phosphate pathway | K00895;K00895;K00895;K00895;K00895 | GO:0030312//external encapsulating structure;GO:0044445//cytosolic part | GO:0008443//phosphofructokinase activity;GO:0032550//purine ribonucleoside binding;GO:0043169//cation binding | GO:0006084//acetyl-CoA metabolic process;GO:0006090//pyruvate metabolic process;GO:0016310//phosphorylation;GO:0016458//gene silencing;GO:0034968//histone lysine methylation | - |
| MSTRG.13315 | antiquewhite4 | 2230.14826120019 | 208.509347923882 | 0.207952719685512 | -0.764746414329568 | -0.379211227695855 | 0.550705272315067 | 0.1670307829493 | 0.217328626197036 | -0.963016965585853 | -0.87409840410392 | -0.602533924009522 | -0.584400715629885 | -0.442739703883782 | 0.839248711562158 | -0.0151289851966742 | 0.404701926417307 | -0.0957708327704047 | 0.0251721883218937 | -0.000213766793618697 | -0.0995826091495116 | -0.0390048931385335 | 0.516623922129397 | 0.00376362443791105 | 0.224098859276324 | 0.0635162519025145 | 0.60385956772302 | 0.497449391572242 | 5.12040056464765e-07 | 0.000200922521851006 | 0.0381265894918165 | 0.0459879172949485 | 0.14948247677354 | 0.000641171872258268 | 0.962779873016627 | 0.191907942179821 | 0.767172576559254 | 0.938105124170692 | 0.999473933313381 | 0.758144973373072 | 0.904205843114152 | 0.00 | 0.11 | 0.13 | 0.92 | 2.09 | 0.45 | 17.62 | 4.18 | 2.88 | 101.52 | 247.51 | 120.04 | -- | glu S.griseus protease inhibitor [Prunus persica] | - | - | - | - | - | - | GO:0006950//response to stress;GO:0010466//negative regulation of peptidase activity | - |
| MSTRG.13789 | antiquewhite4 | 1751.157652632 | 106.808841651936 | 0.1504977774664 | 0.74932061542811 | 0.378456940723274 | -0.472200483175366 | -0.115684117180504 | -0.113101125978925 | 0.843738171131703 | 0.842487589276766 | 0.526710523320982 | 0.622230160608032 | 0.426897222686915 | -0.844868475572642 | -0.168871237099333 | -0.494024580691575 | -0.0408353975397036 | 0.0118829667069112 | -0.0443193981663666 | 0.0974801794206584 | 0.297175684227251 | 0.640593660328039 | 0.00502609936473553 | 0.225097744762832 | 0.121133881402045 | 0.720327783789015 | 0.72635810473506 | 0.000560948708133693 | 0.000582465641236298 | 0.0785102057258242 | 0.0307270489816929 | 0.16634339972378 | 0.000542036086536081 | 0.599820145342656 | 0.102581842529591 | 0.899729738026525 | 0.970762266638048 | 0.891217867816709 | 0.763120897886084 | 0.348222577985019 | 0.86 | 1.15 | 1.30 | 1.01 | 0.95 | 1.23 | 0.78 | 0.70 | 1.42 | 0.72 | 0.27 | 0.21 | -- | PREDICTED: transposase, partial [Prunus dulcis] | - | - | - | - | - | - | - | - |
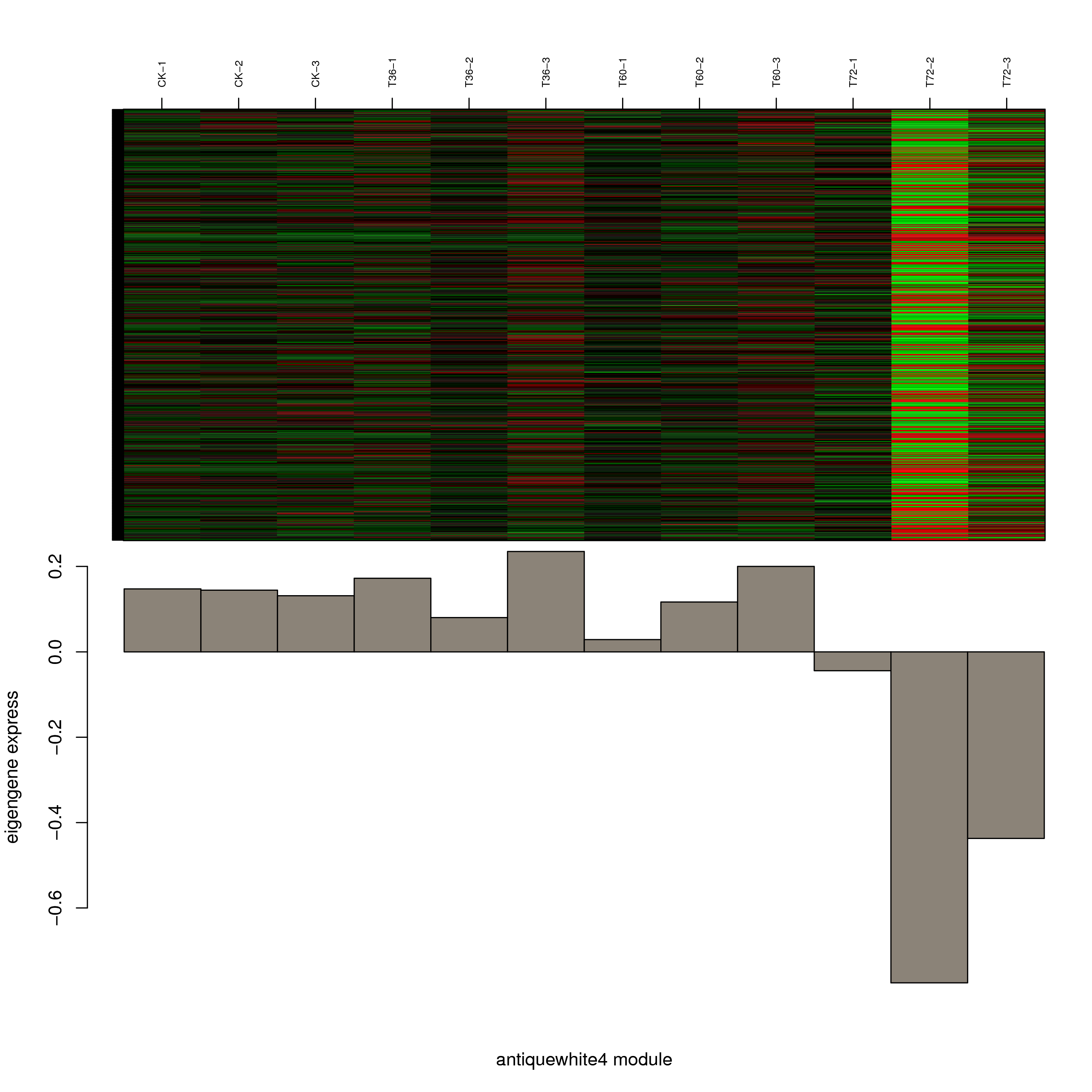
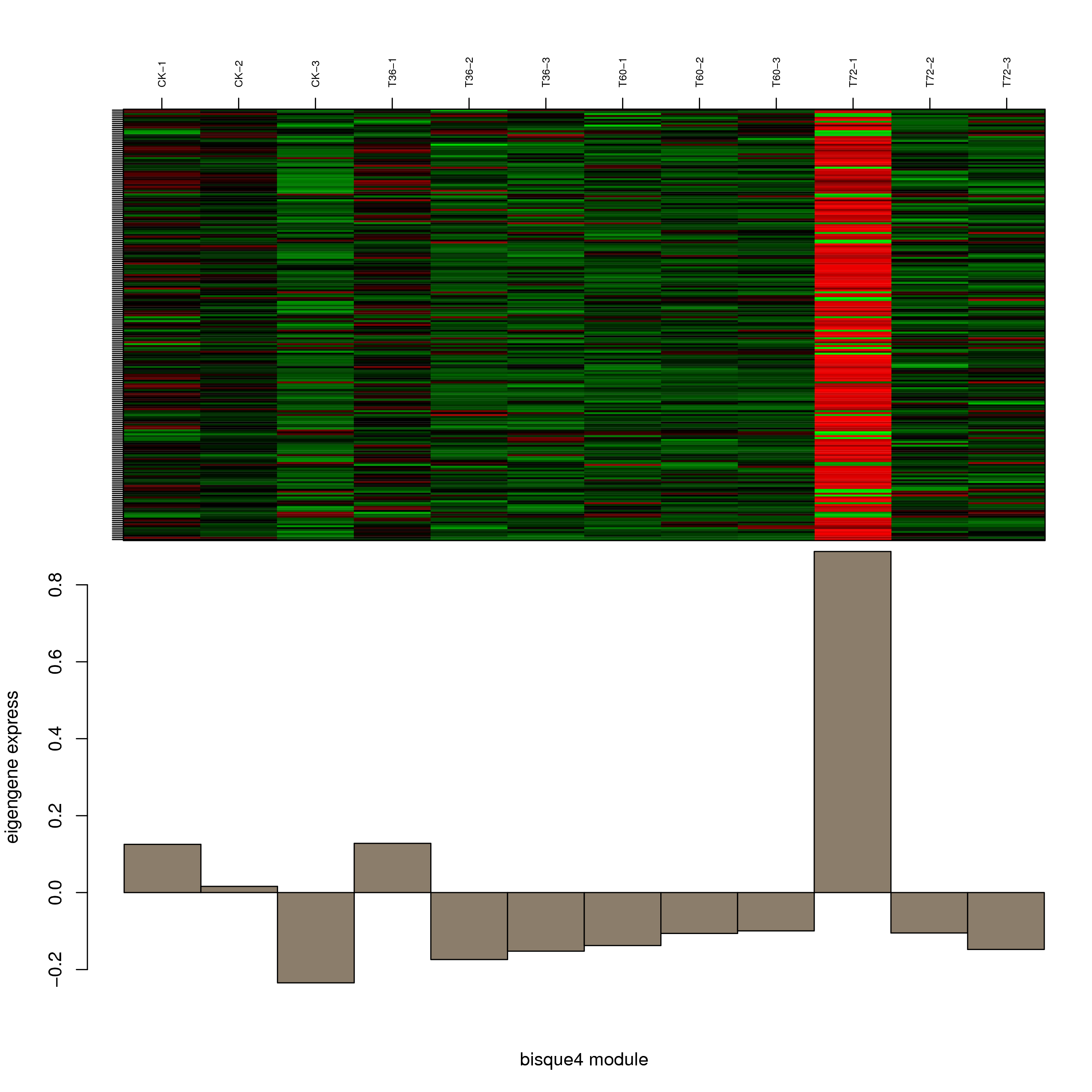
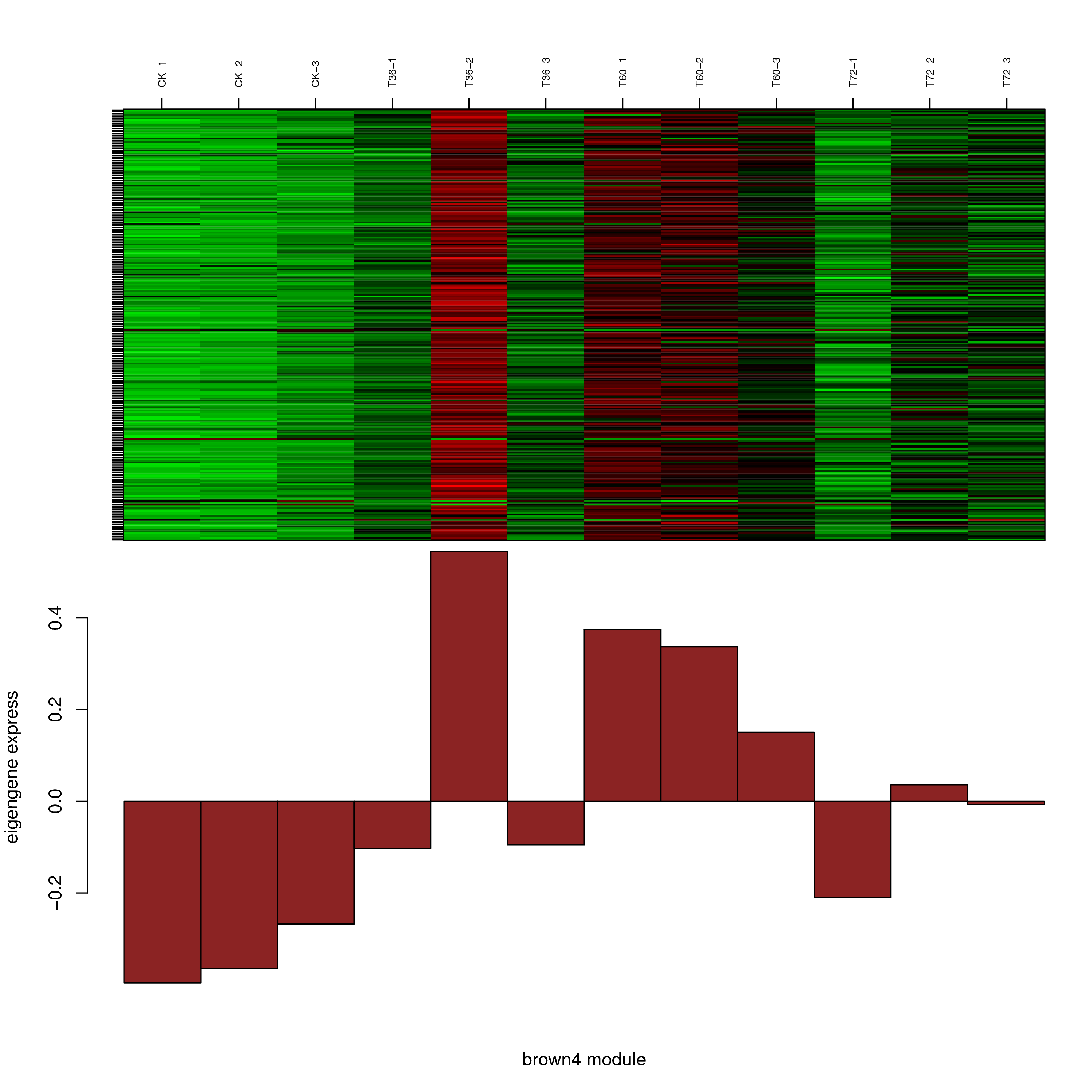
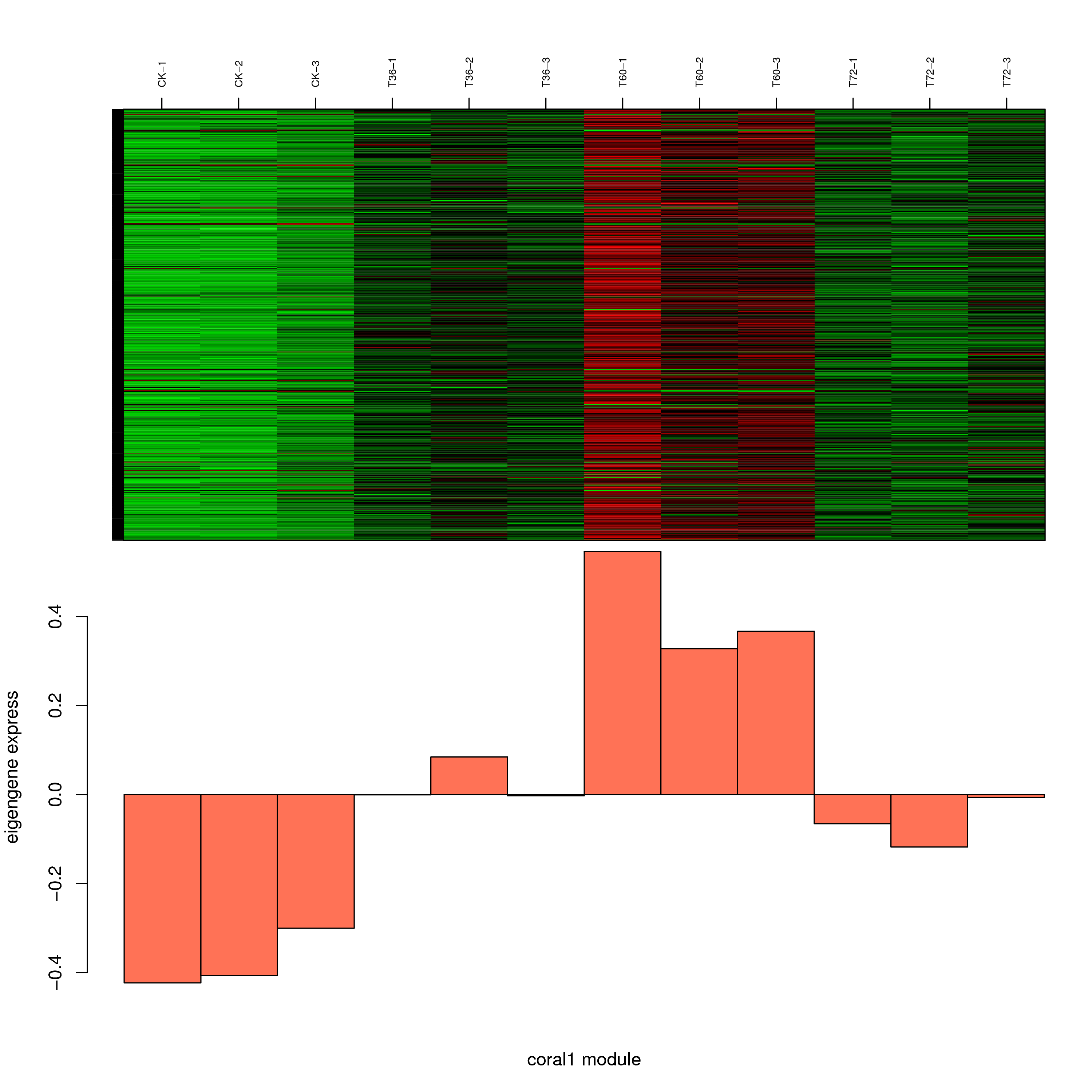
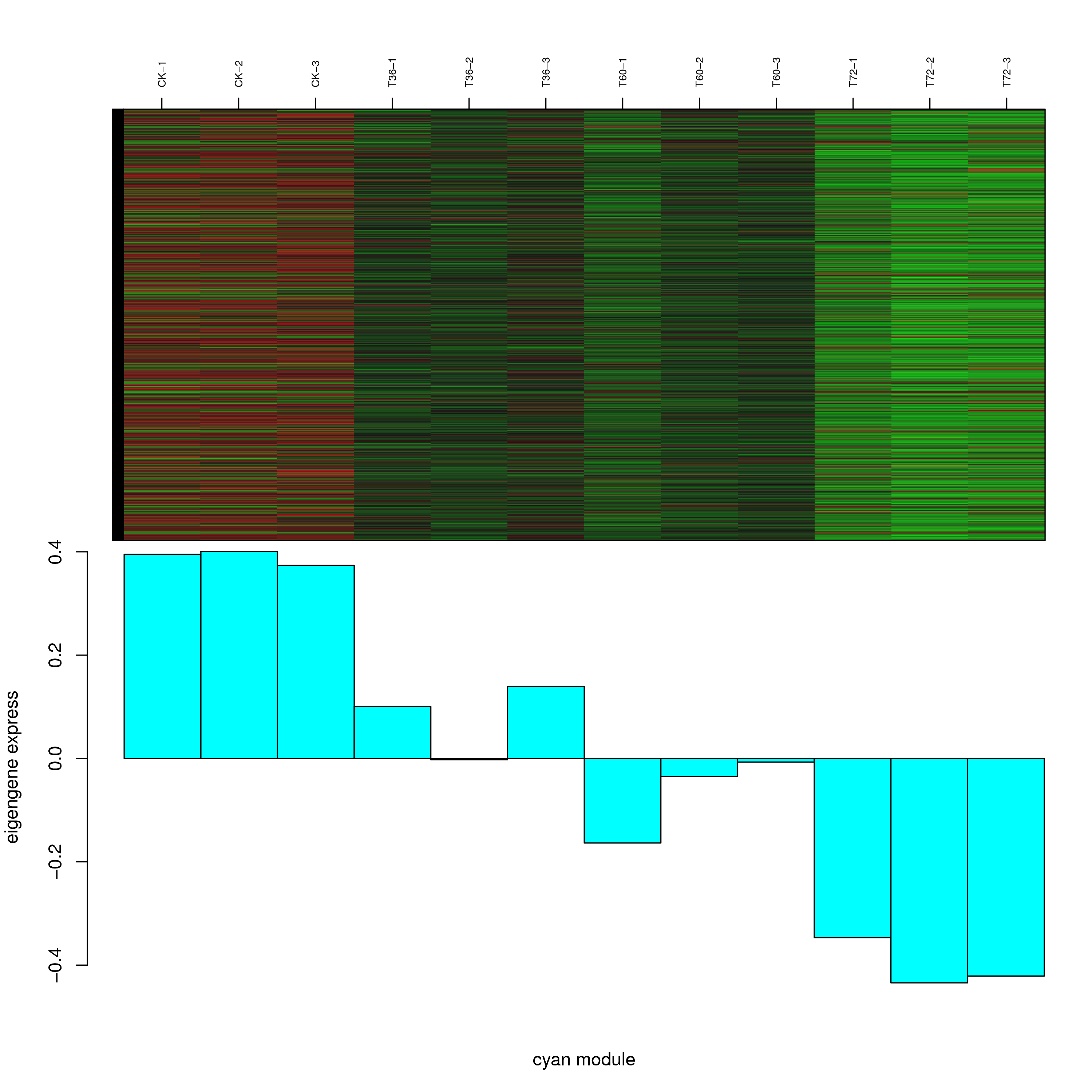
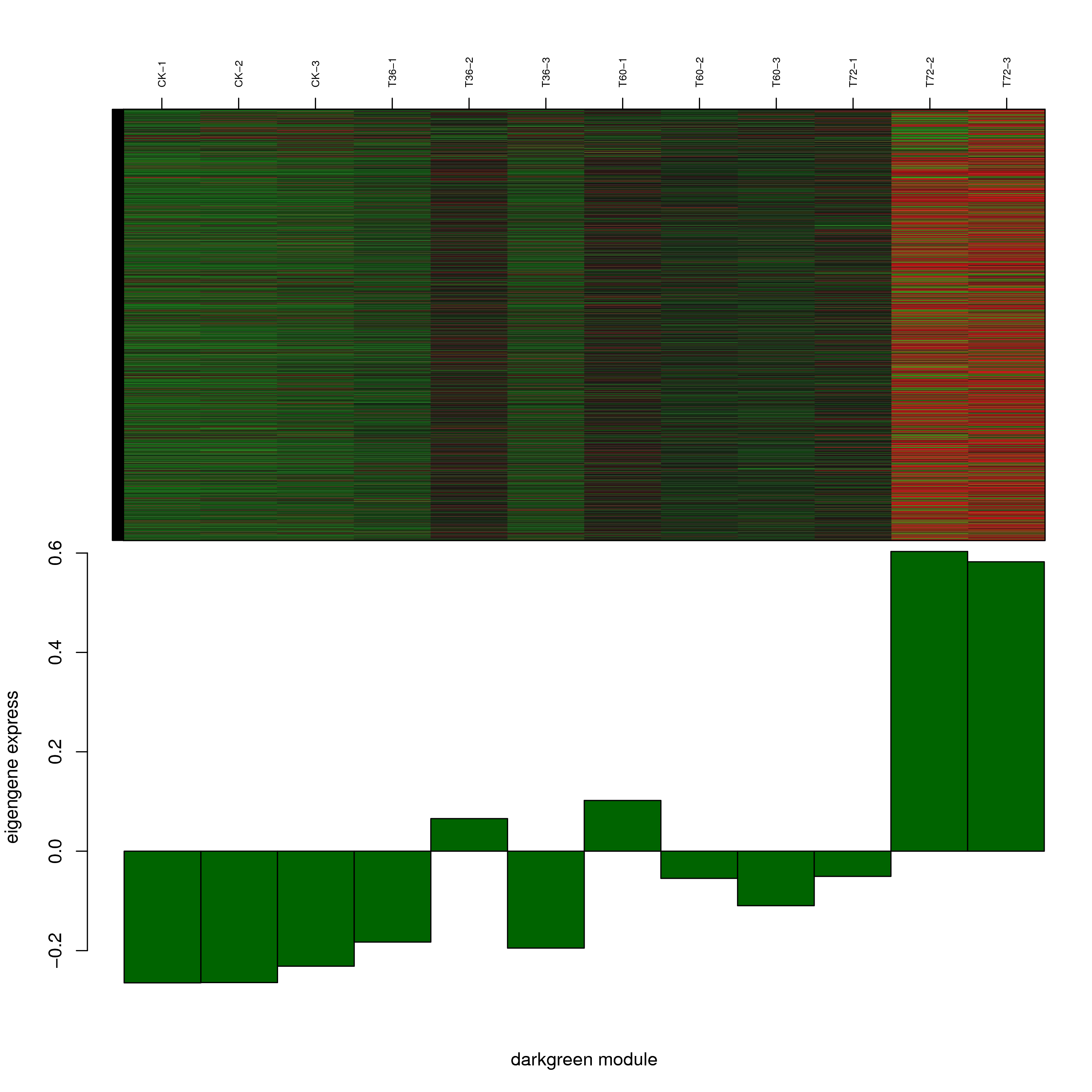
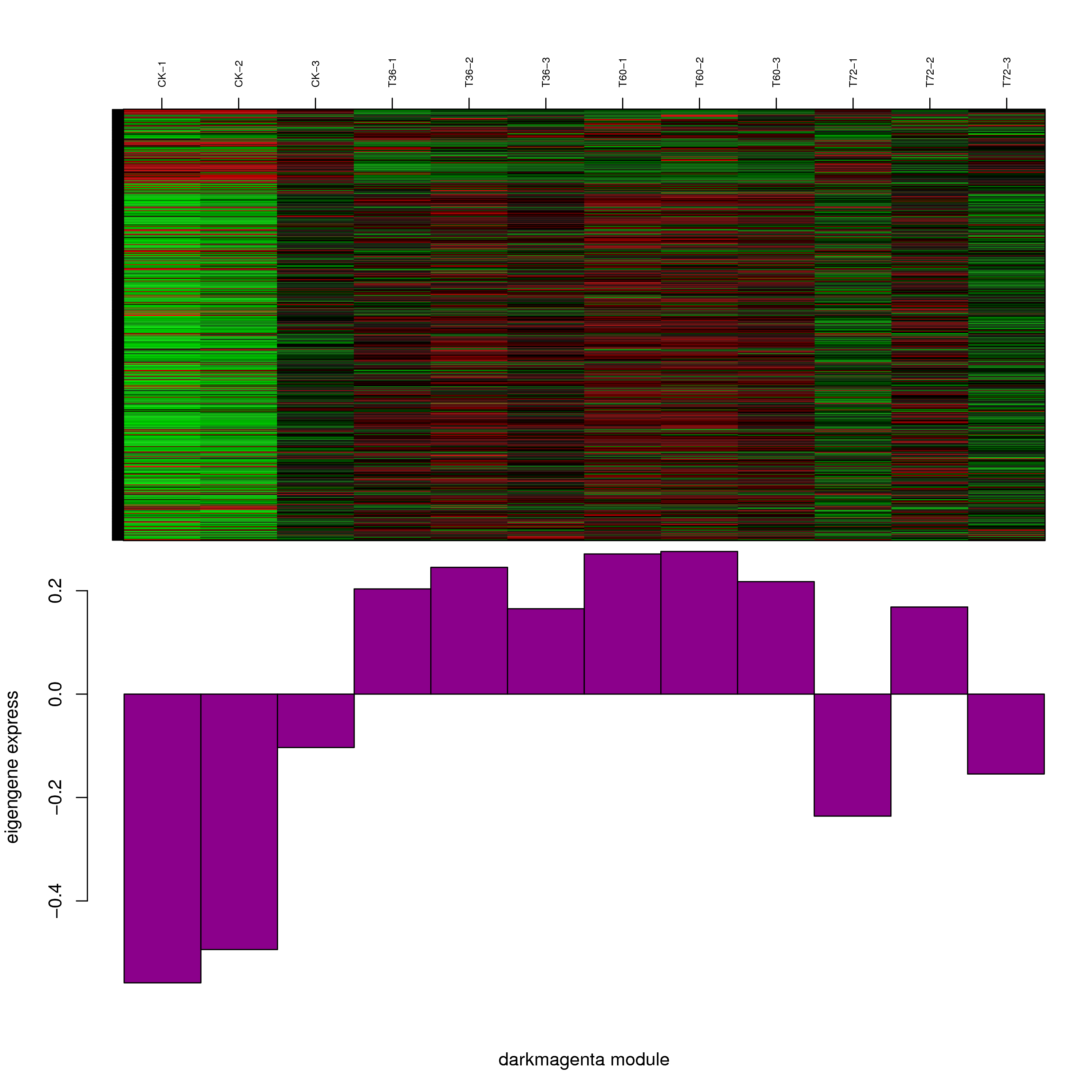
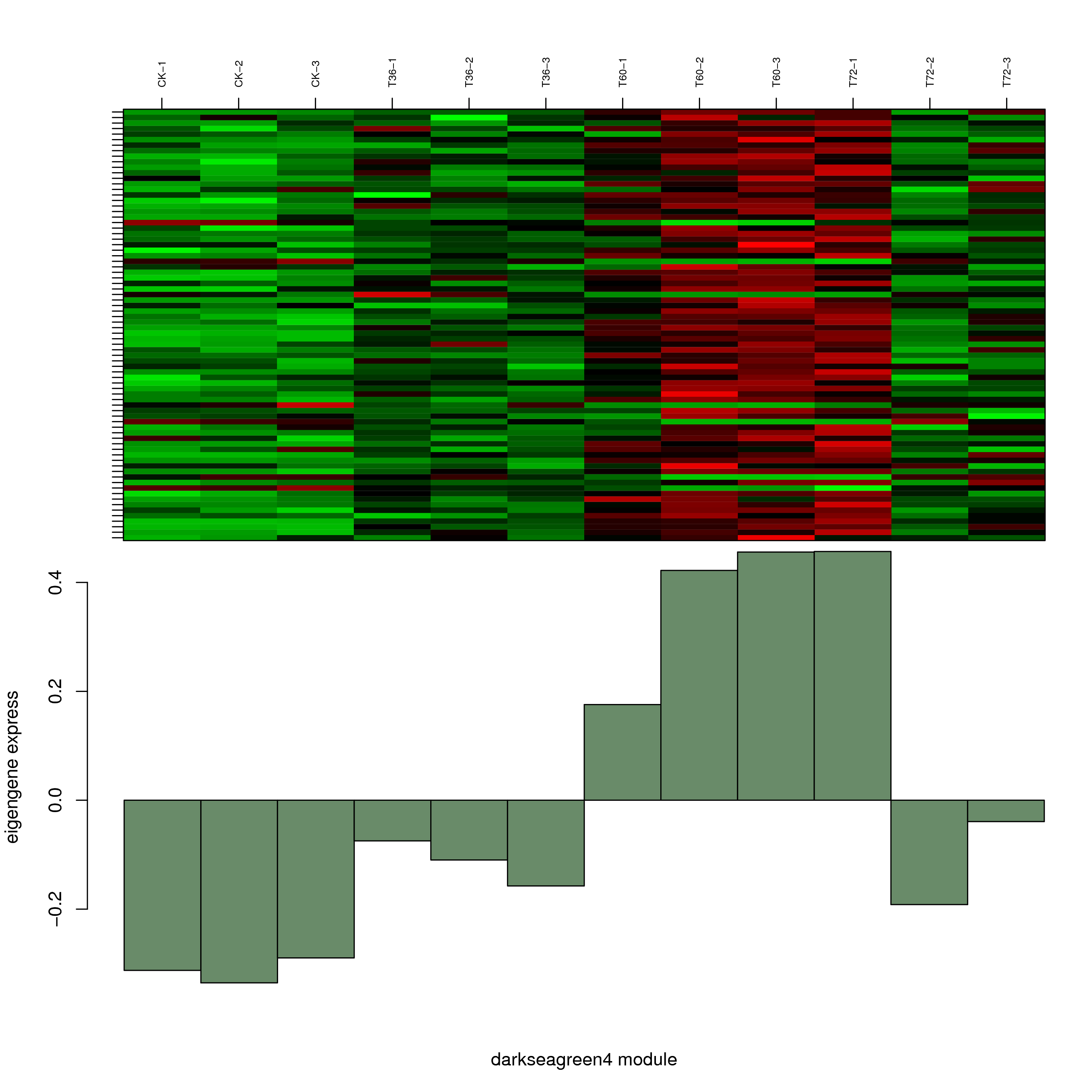
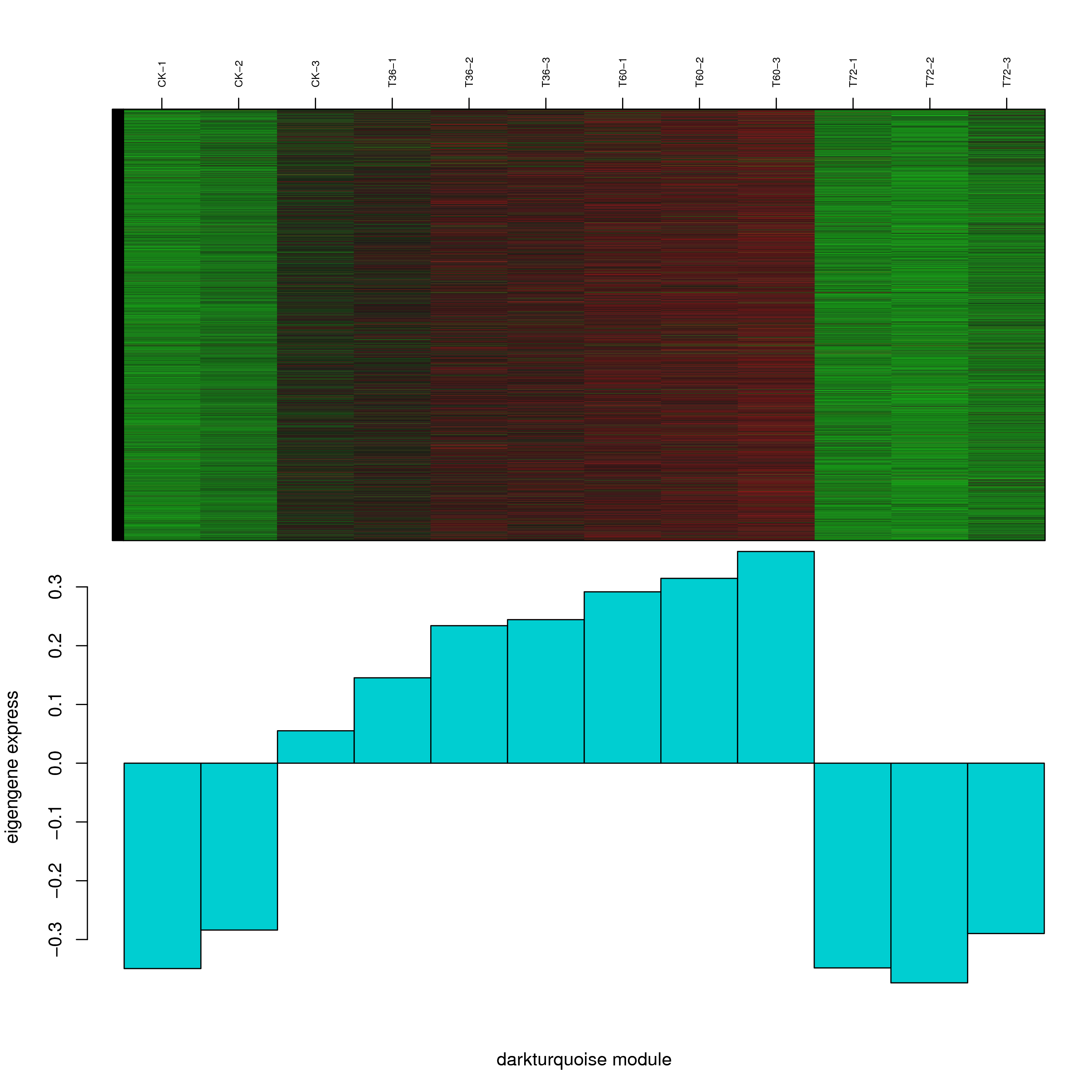
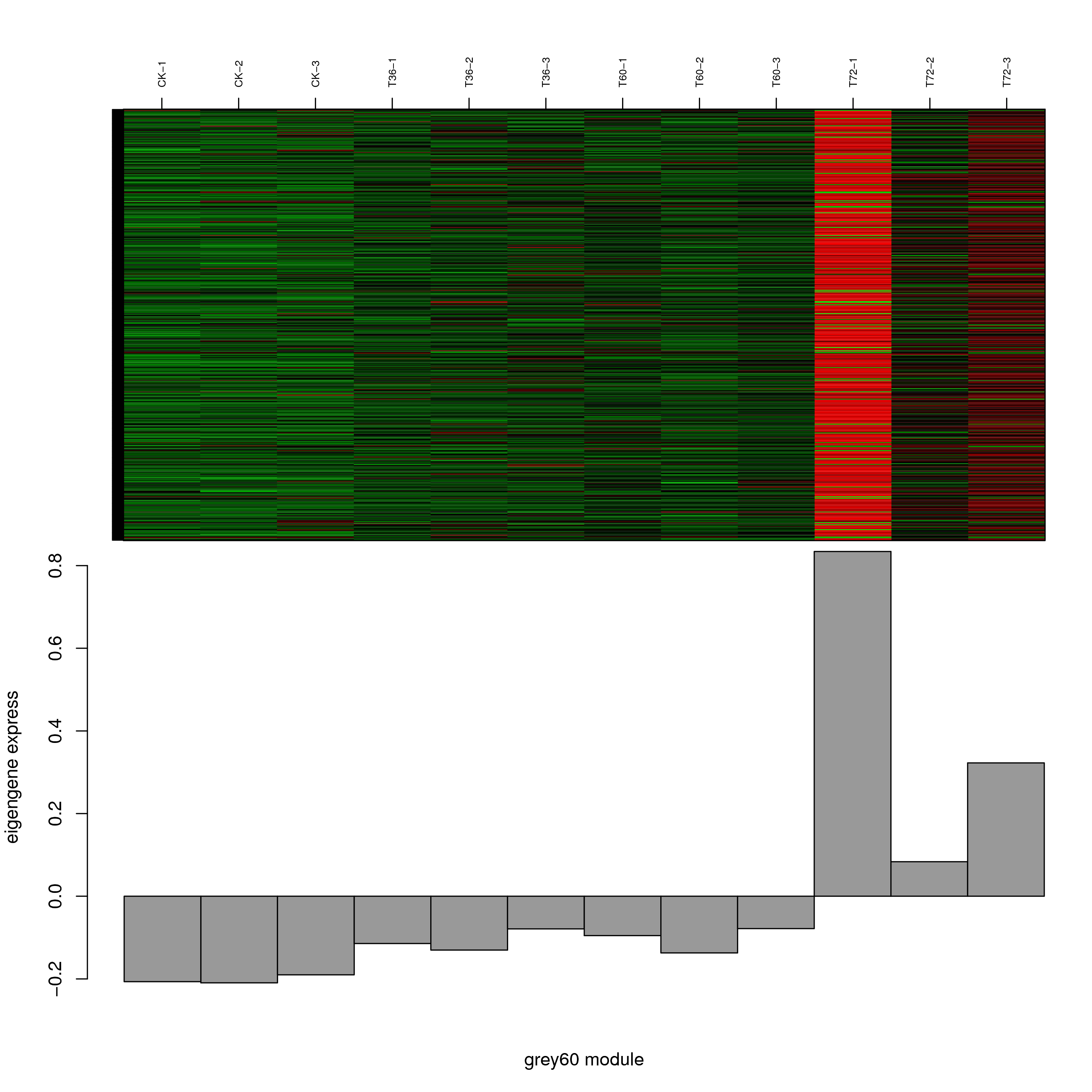
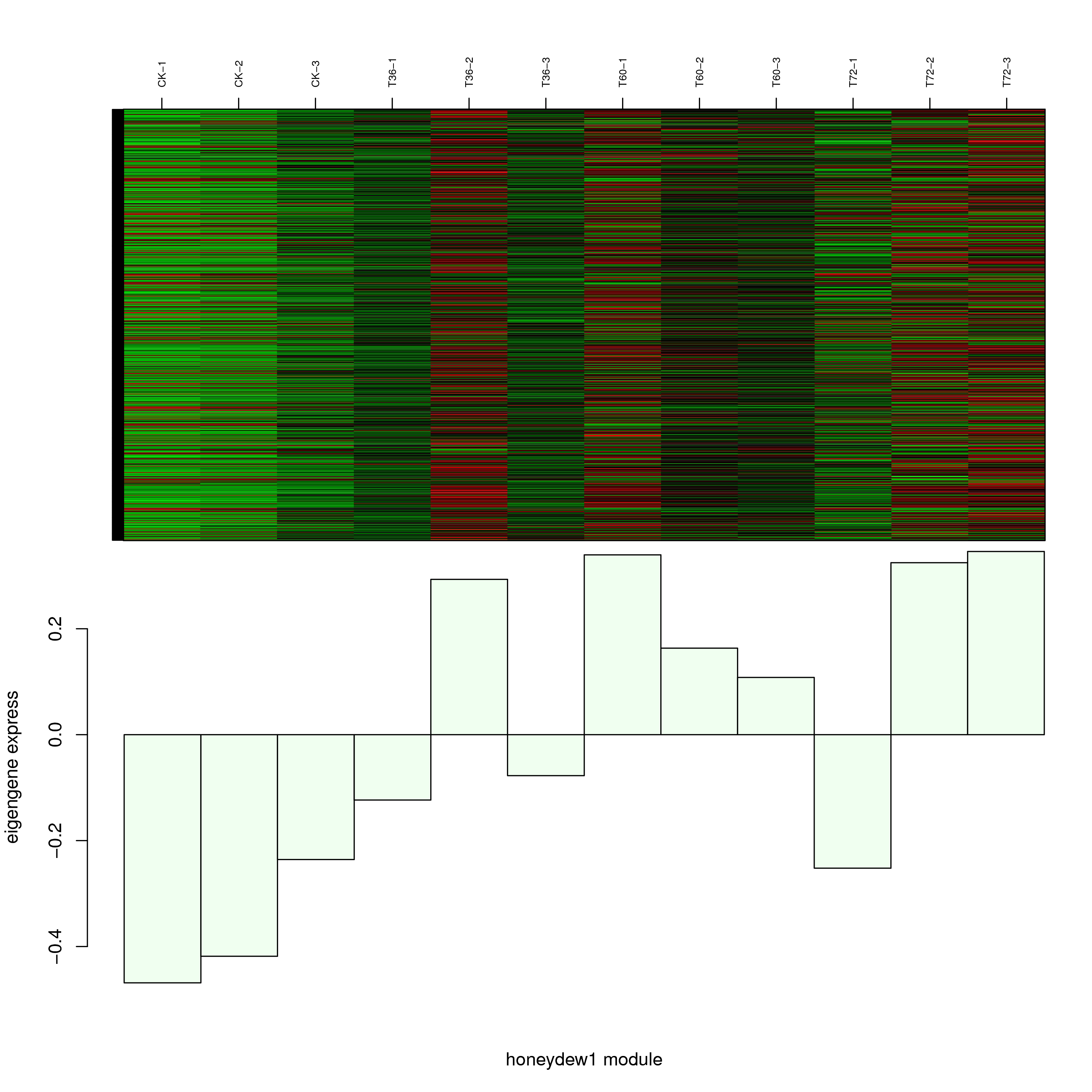
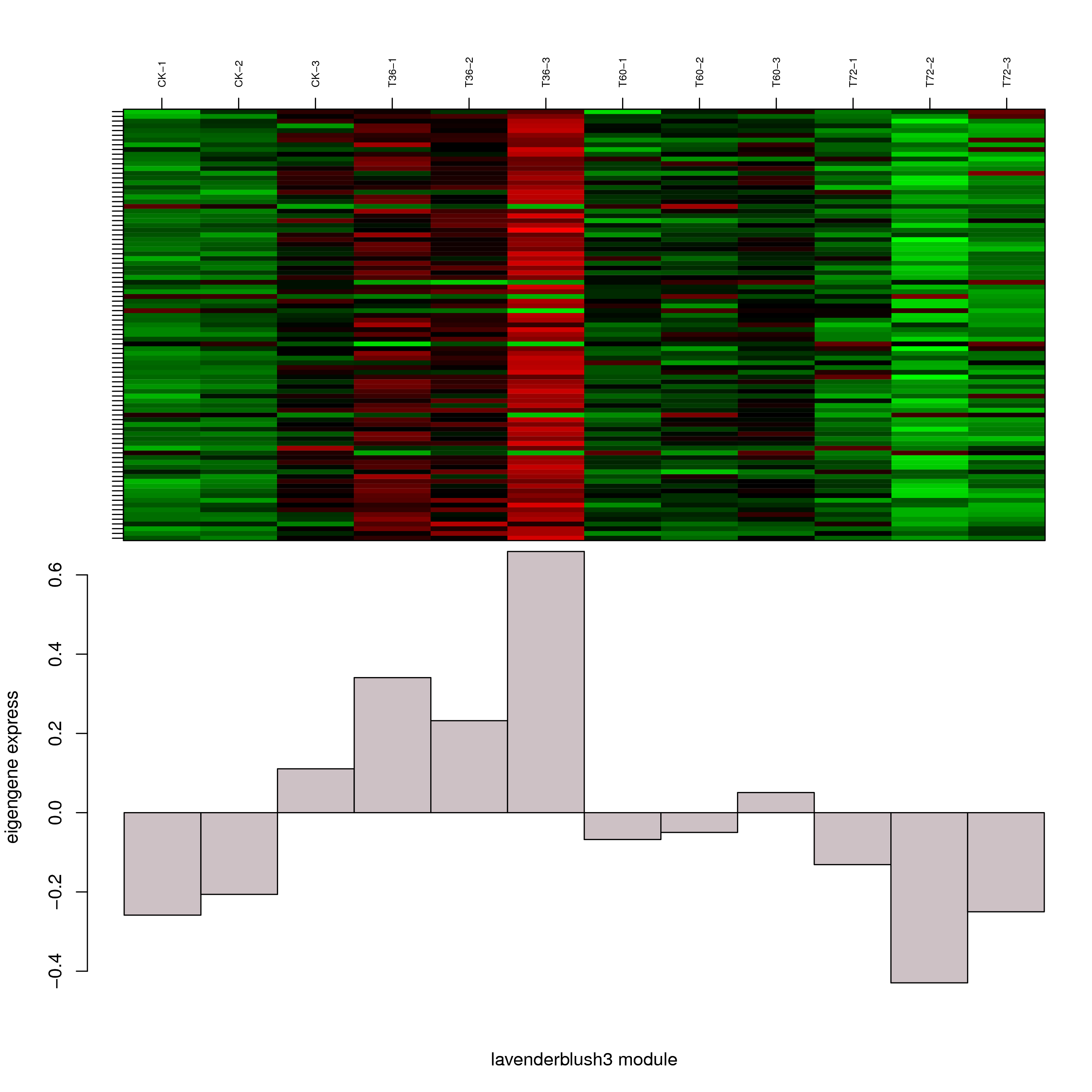
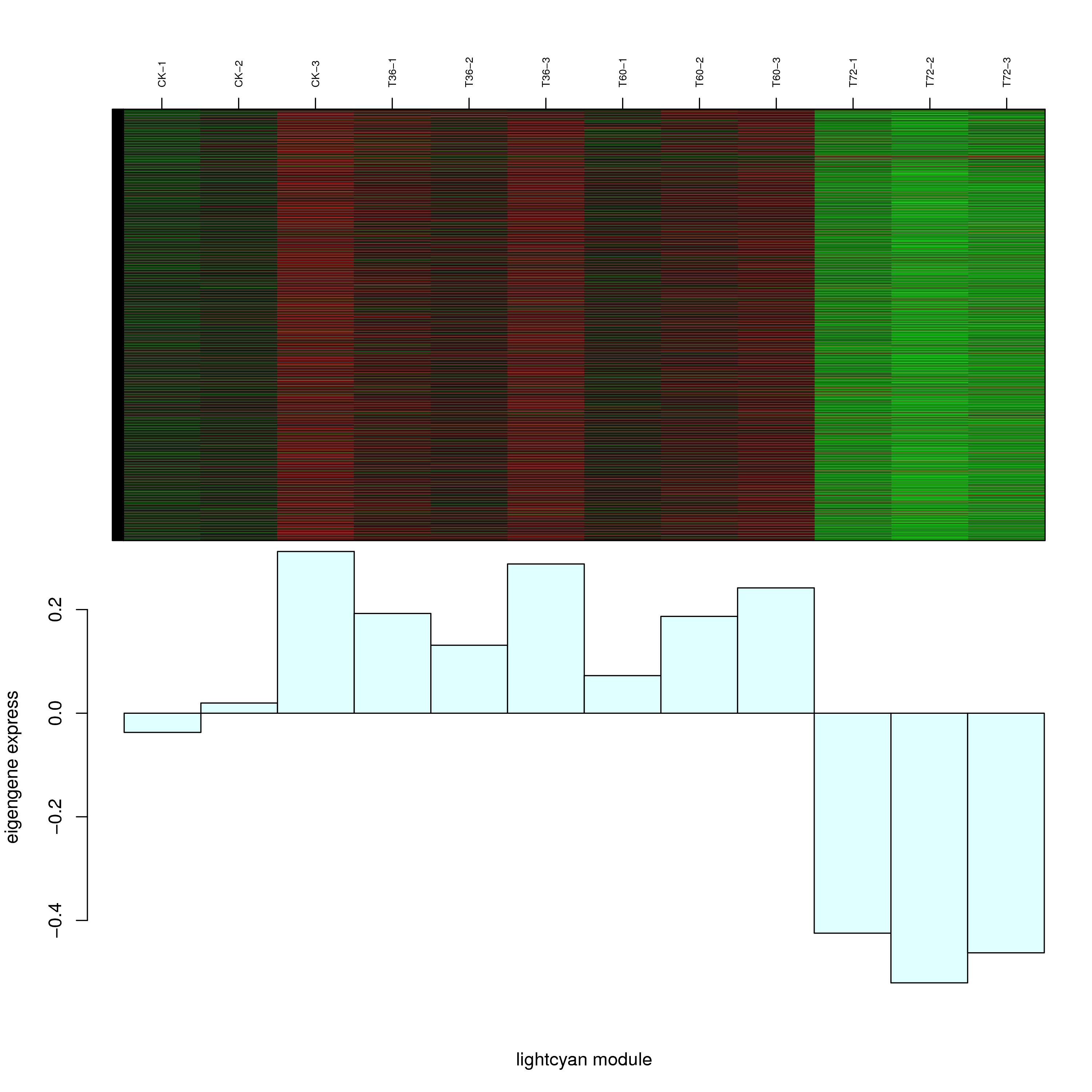
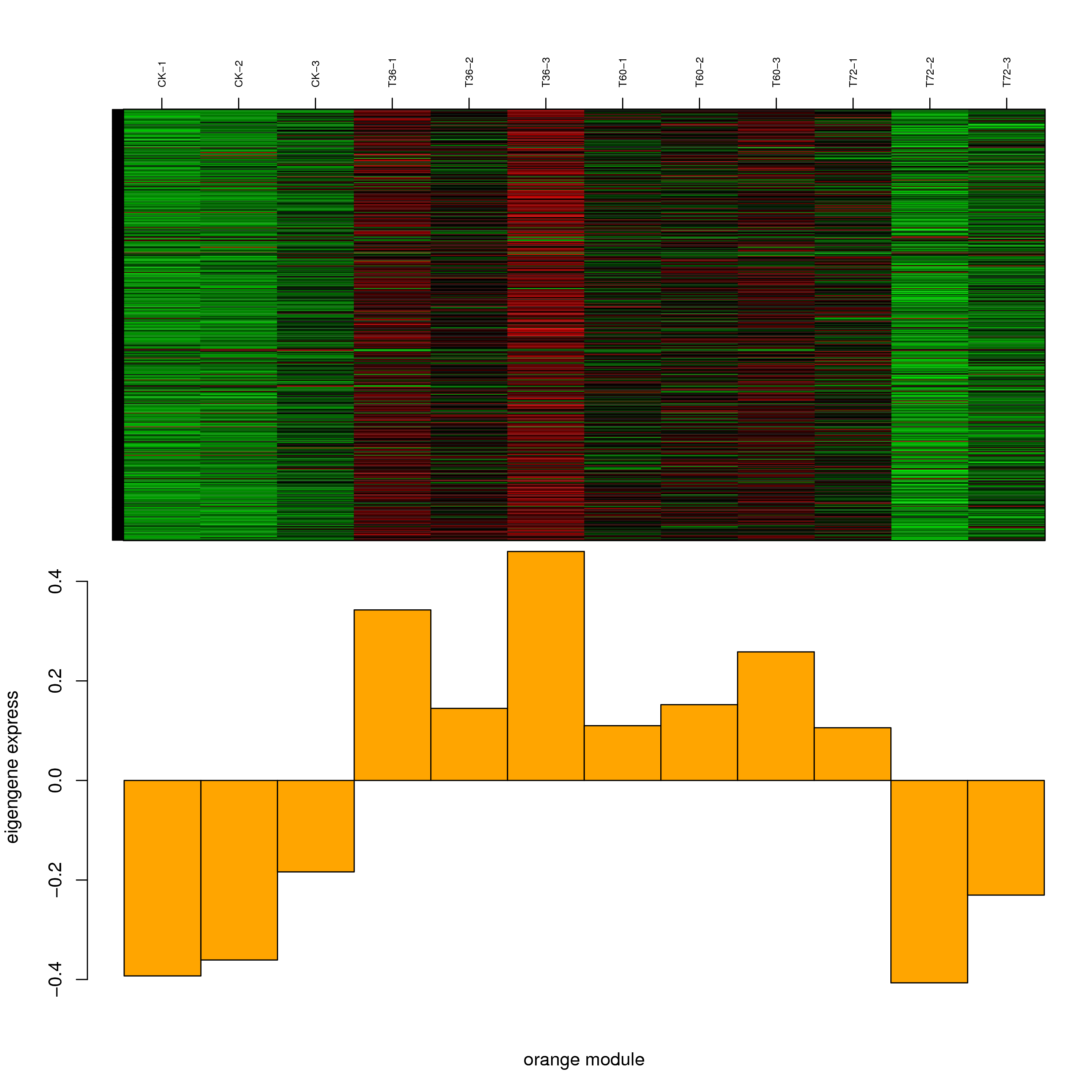
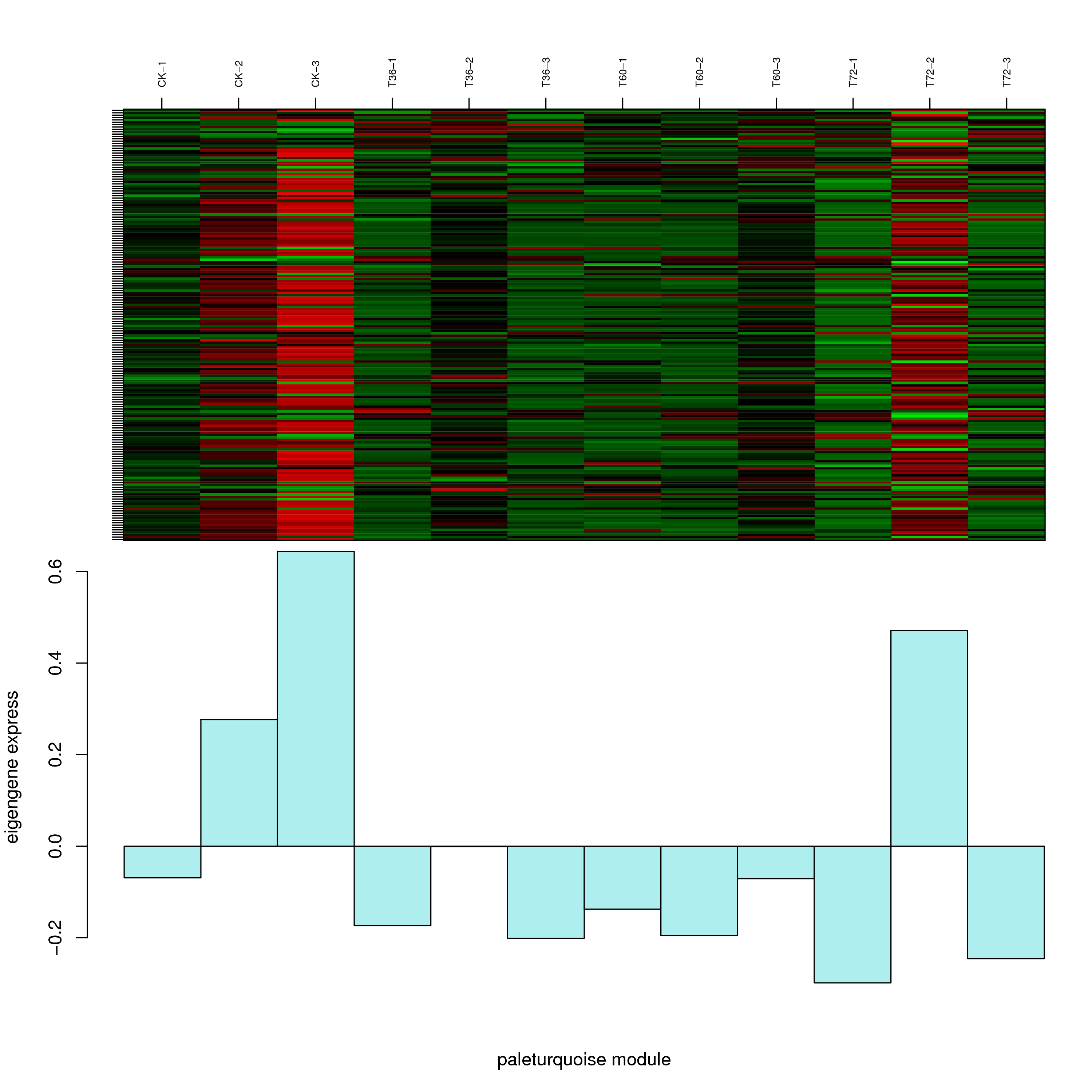
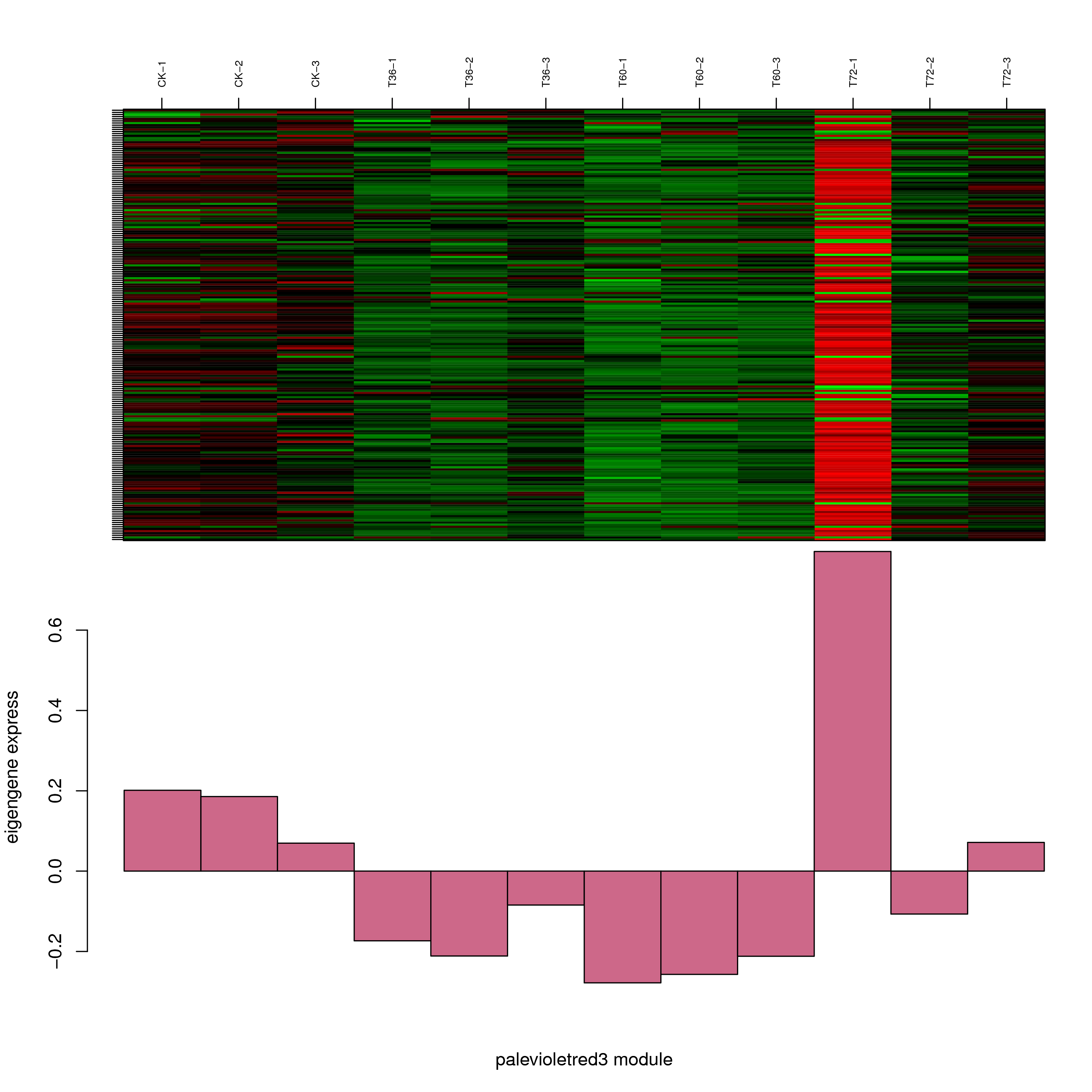
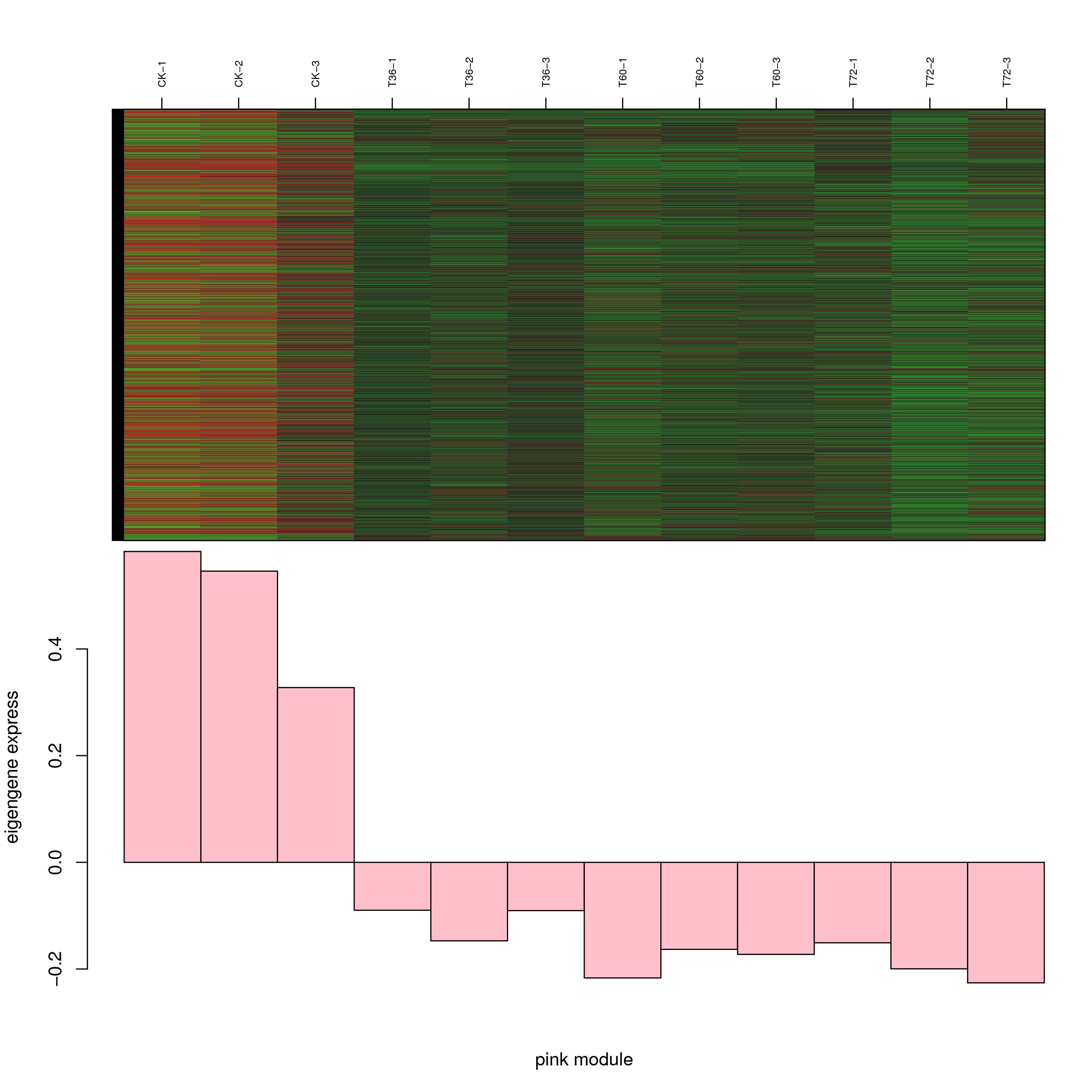
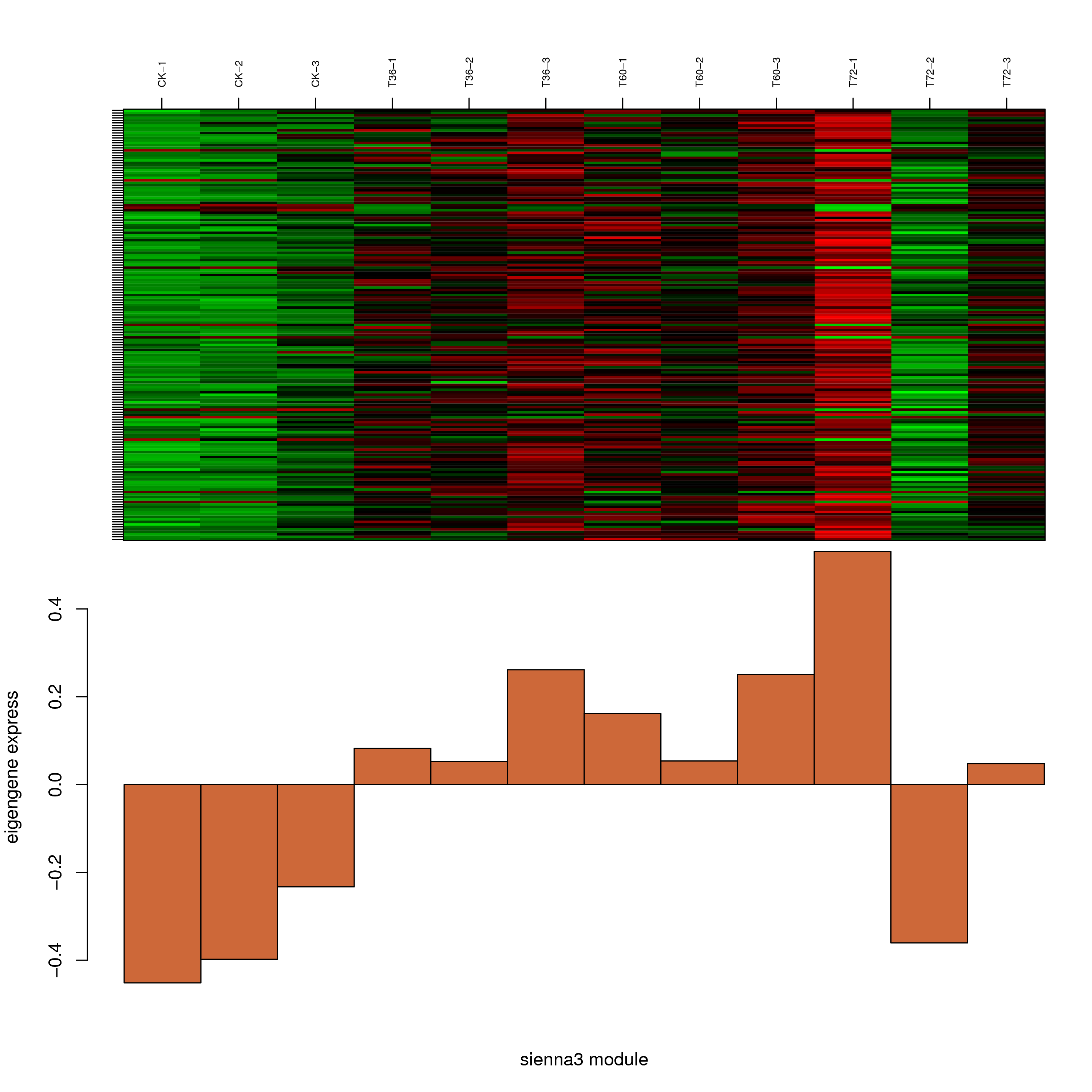
6.2 模块基因表达模式
将每个模块包含的各个基因的表达模式用热图展示,并用柱状图呈现模块特征值在不同样本的变化(相当于模块表达模式)。图中,上图为模块中基因在不同样本中的表达量热图,红色为上调,绿色为下调;下图为在不同样本中的模块特征值。
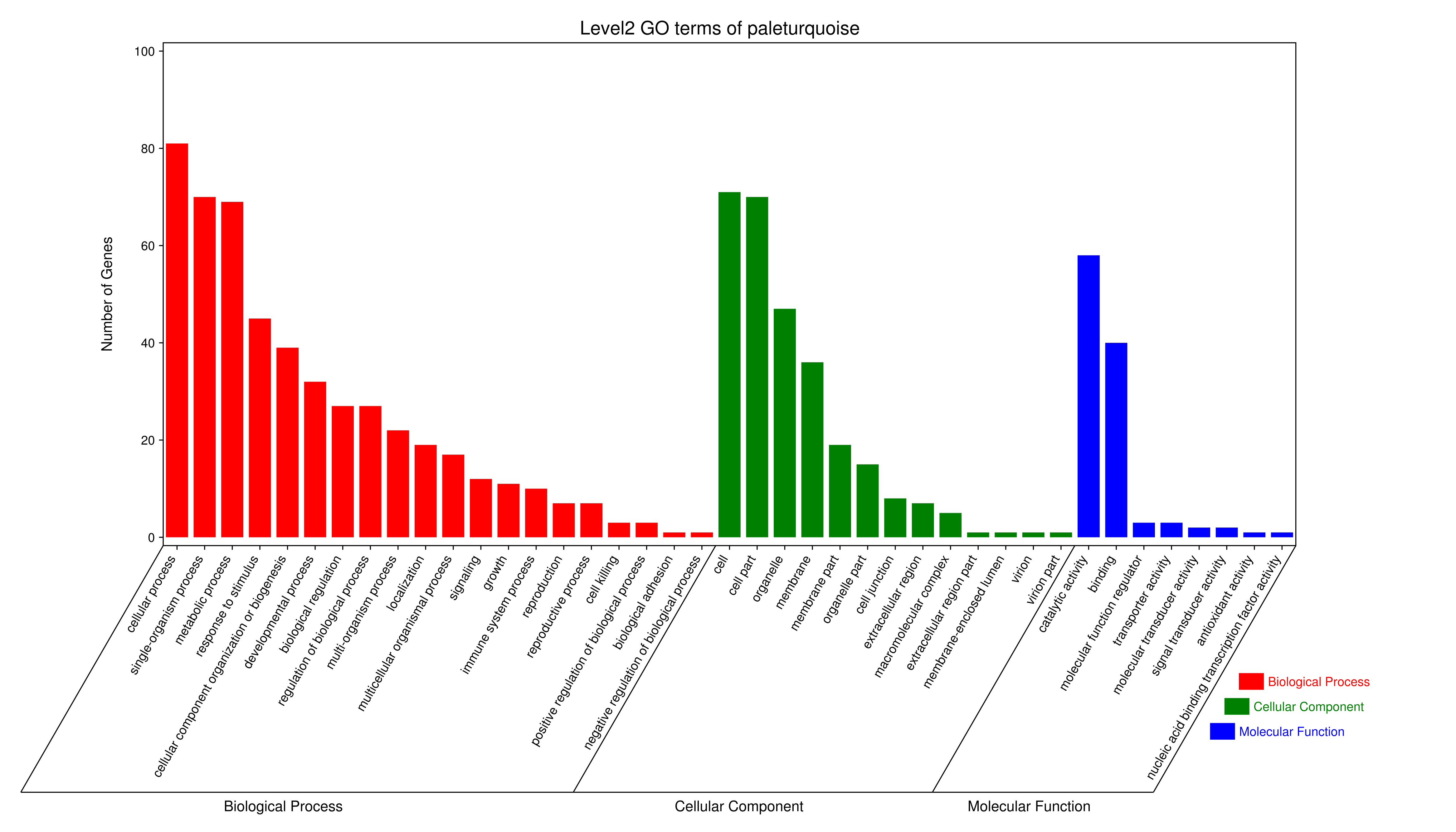
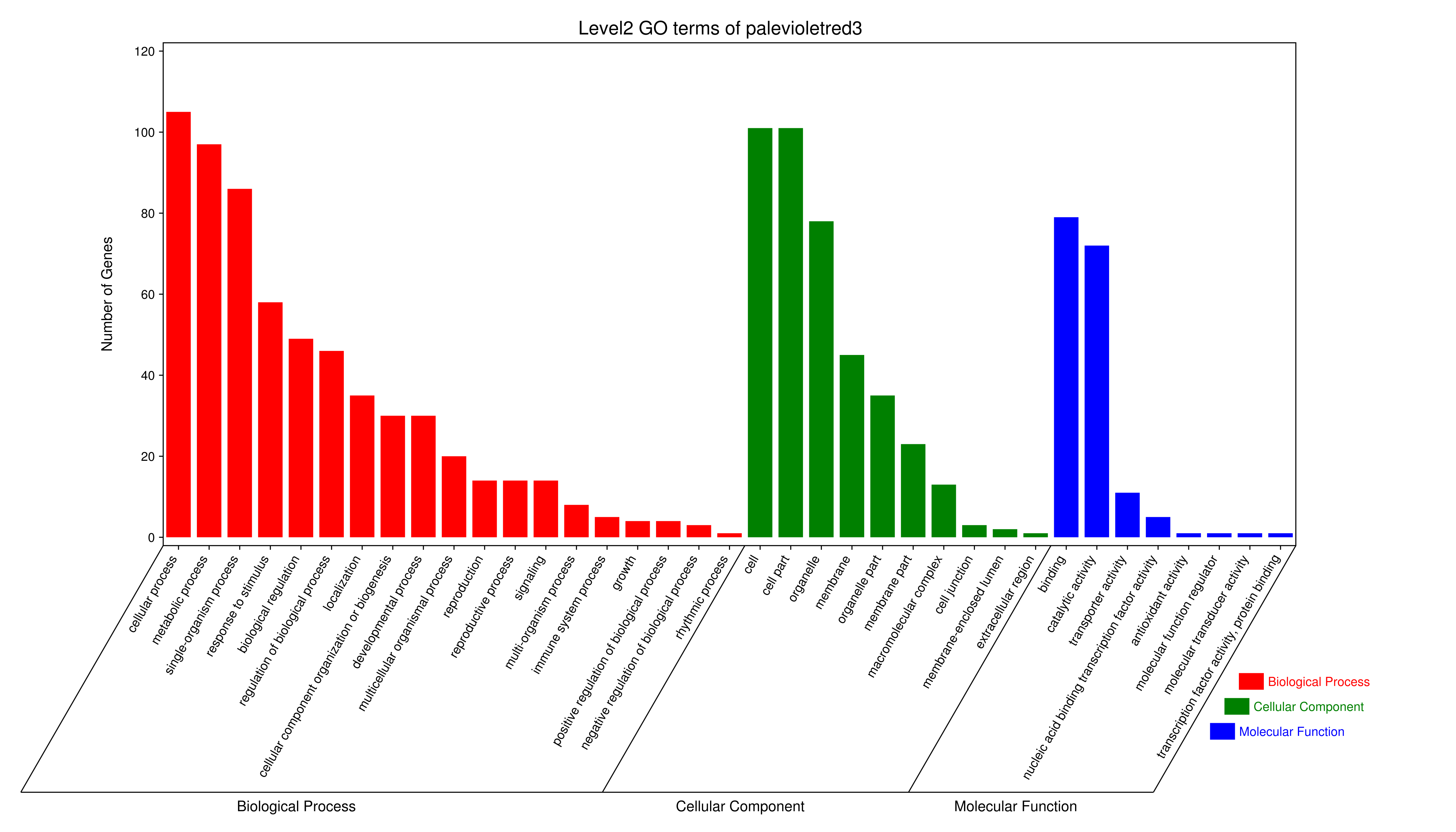
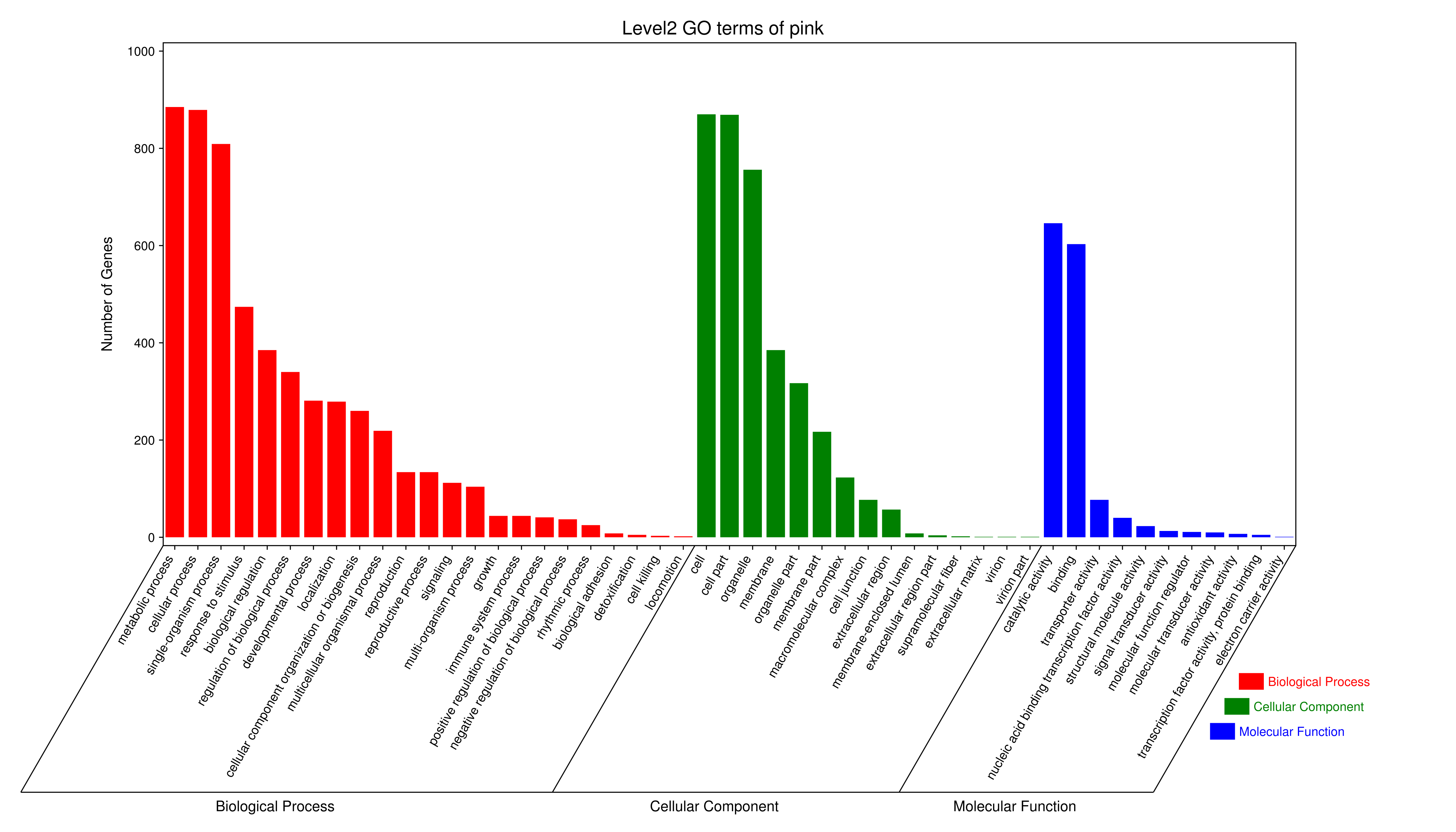
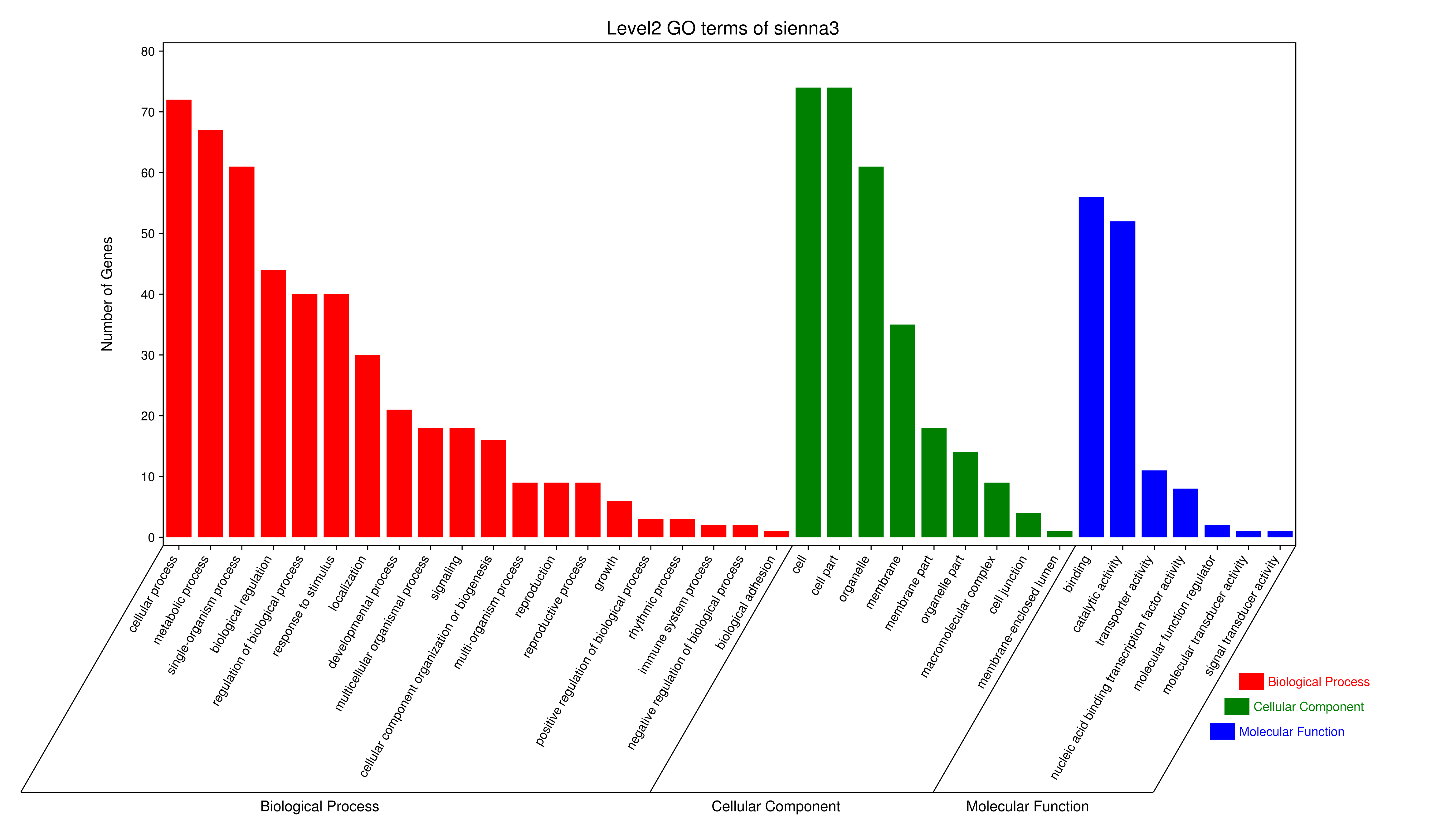
- antiquewhite4
- bisque4
- brown4
- coral1
- cyan
- darkgreen
- darkmagenta
- darkseagreen4
- darkturquoise
- grey60
- honeydew1
- lavenderblush3
- lightcyan
- orange
- paleturquoise
- palevioletred3
- pink
- sienna3
6.3 各模块网络信息(可用于cytoscape作图)
我们提供各模块节点文件和边界文件,可用于导入cytoscape软件[5],构建基因共表达网络图。也可挑选感兴趣的一部分基因(如连通性较高的基因),构建基因共表达网络图。基因调控关系网络图能帮助我们获取在调控关系网络中处于枢纽位置的核心基因(hub gene),并且可以利用已知基因的功能预测未知基因功能(直线相连的基因功能潜在相关)。我们还提供TF因子的信息,在绘制调控网络图时,可以将TF因子在图中标注出来。
注:依据系统配置及浏览器不同,如果关系对数量过多该图可能不能正常加载,请使用桌面版Cytoscape软件
下图展示的为示例图,可以根据个人喜好使用桌面版Cytoscape软件调整图形。
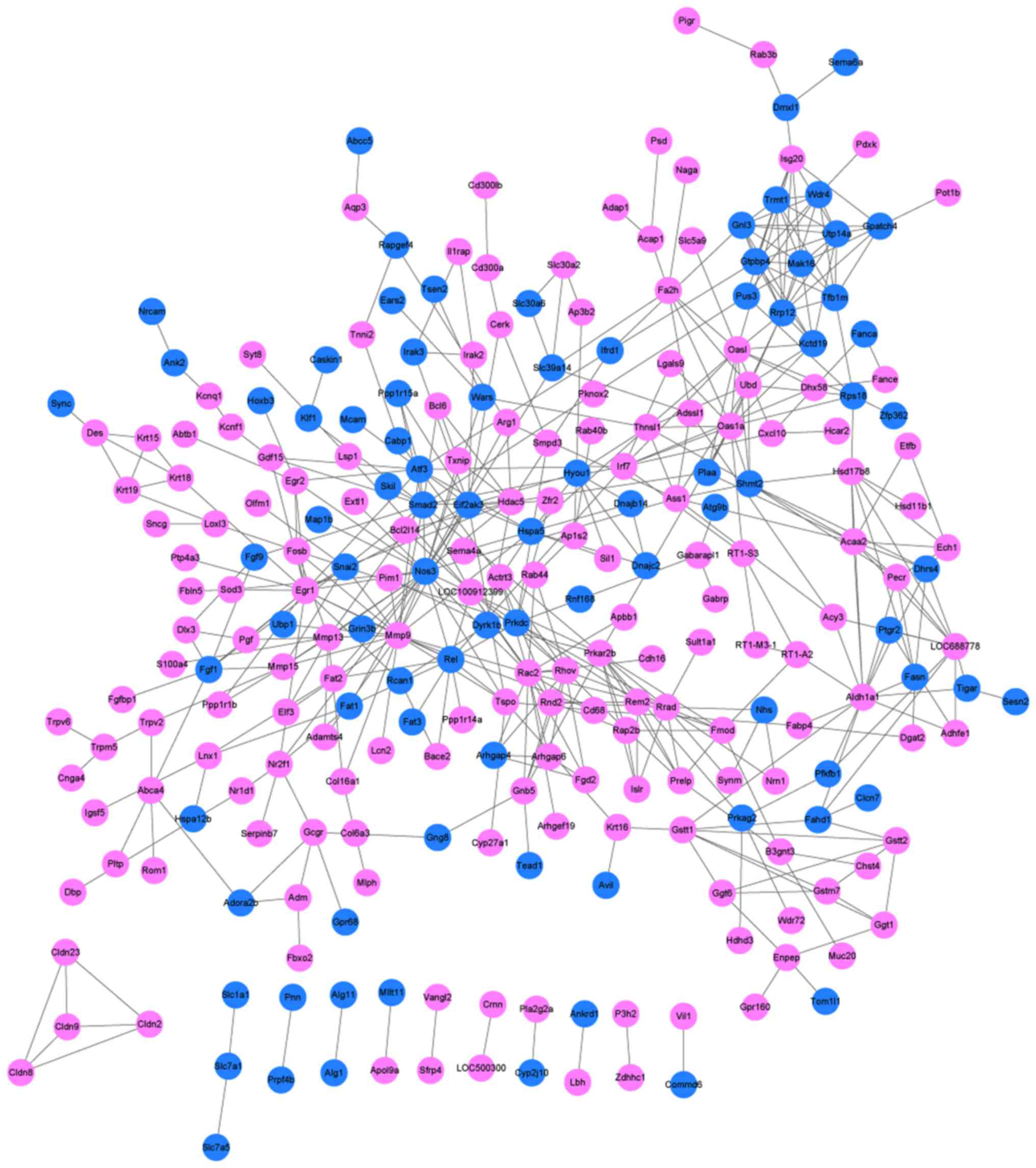 |
| Fig 6-3-1 网络图例图 |
8 参考文献
[1] Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis[J]. Statistical applications in genetics and molecular biology, 2005, 4(1). 返回
[2] Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis[J]. BMC bioinformatics, 2008, 9(1): 559. 返回
[3] Jin J, Zhang H, Kong L, et al. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors[J]. Nucleic acids research, 2013, 42(D1): D1182-D1187. 返回
[4] Zhang H M, Chen H, Liu W, et al. AnimalTFDB: a comprehensive animal transcription factor database[J]. Nucleic acids research, 2011, 40(D1): D144-D149. 返回
[5] Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks[J]. Genome research, 2003, 13(11): 2498-2504. 返回
[6] Ashburner M, Ball C A, Blake J A, et al. Gene ontology: tool for the unification of biology[J]. Nature genetics, 2000, 25(1): 25. 返回
[7] Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes[J]. Nucleic acids research, 2000, 28(1): 27-30. 返回
9 附录
9.1 分析方法英文文档
9.2 结果目录结构
result
├── 1.Filter 数据过滤
│ |-- removeGene.xls 过滤掉的基因列表
│ |-- removeSample.xls 过滤掉的样本列表
├── 2.module_construction 模块划分
│ |-- 1.sampleClustering.pdf/png 样本层次聚类树图
│ |-- 2.softPower.pdf/png Power值曲线图
│ |-- 3.eigengeneClustering.pdf/png 模块特征值聚类图
│ |-- 4.ModuleTree.pdf/png 模块层次聚类图
│ |-- 4.netcolor2gene.xls 基因-模块对应关系列表
│ |-- 5.module_gene.xls 各模块基因统计表
│ |-- 5.module_gene.pdf/png 各模块基因数柱状图
├── 3.basic_info 模块相关信息
│ |-- 1.ModuleModuleMembership.xls/pdf/png 模块-模块相关性结果
│ |-- 2.geneModuleMembership.xls/pdf/png 模块-基因相关性结果
│ |-- 3.SampleExpressionPattern.xls/pdf/png 样本表达模式分析
│ |-- 4.hubGene.xls Hub基因相关结果
│ |-- 5.ModuleTraitRelation.xls/pdf/png 性状关联相关性分析
│ |-- TF.xls TF相关性结果
│ |-- TFmodule.xls/pdf/png TF因子统计结果
│ |-- MM-GS-*-Scatter.png 某性状最正相关和最负相关模块的MM 和 GS相关性散点图
│ |-- K.in-GS-*-Scatter.png 某性状最正相关和最负相关模块的K.in 和 GS相关性散点图
│ |-- moduleGS-*-histogram.png 各模块与某性状的GS的柱状图
├── 4.modules 各模块结果
│ |-- all.glist.xls 模块基因总表
│ |-- *.glist.xls 各模块基因信息表格
│ |-- *_Express.pdf/png 各模块基因表达模式热图
│ |-- cytoscape 网络图所需文件目录
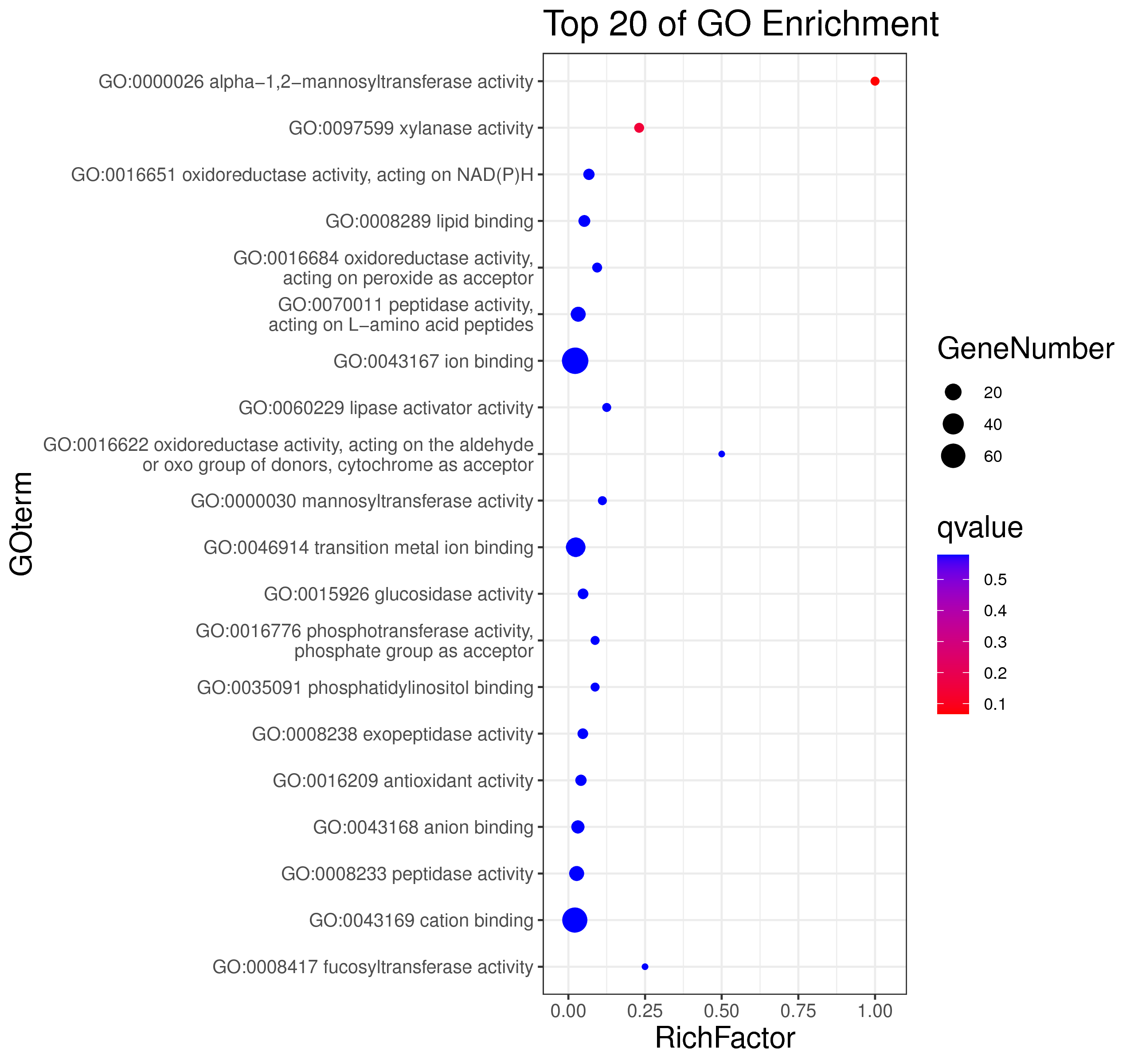
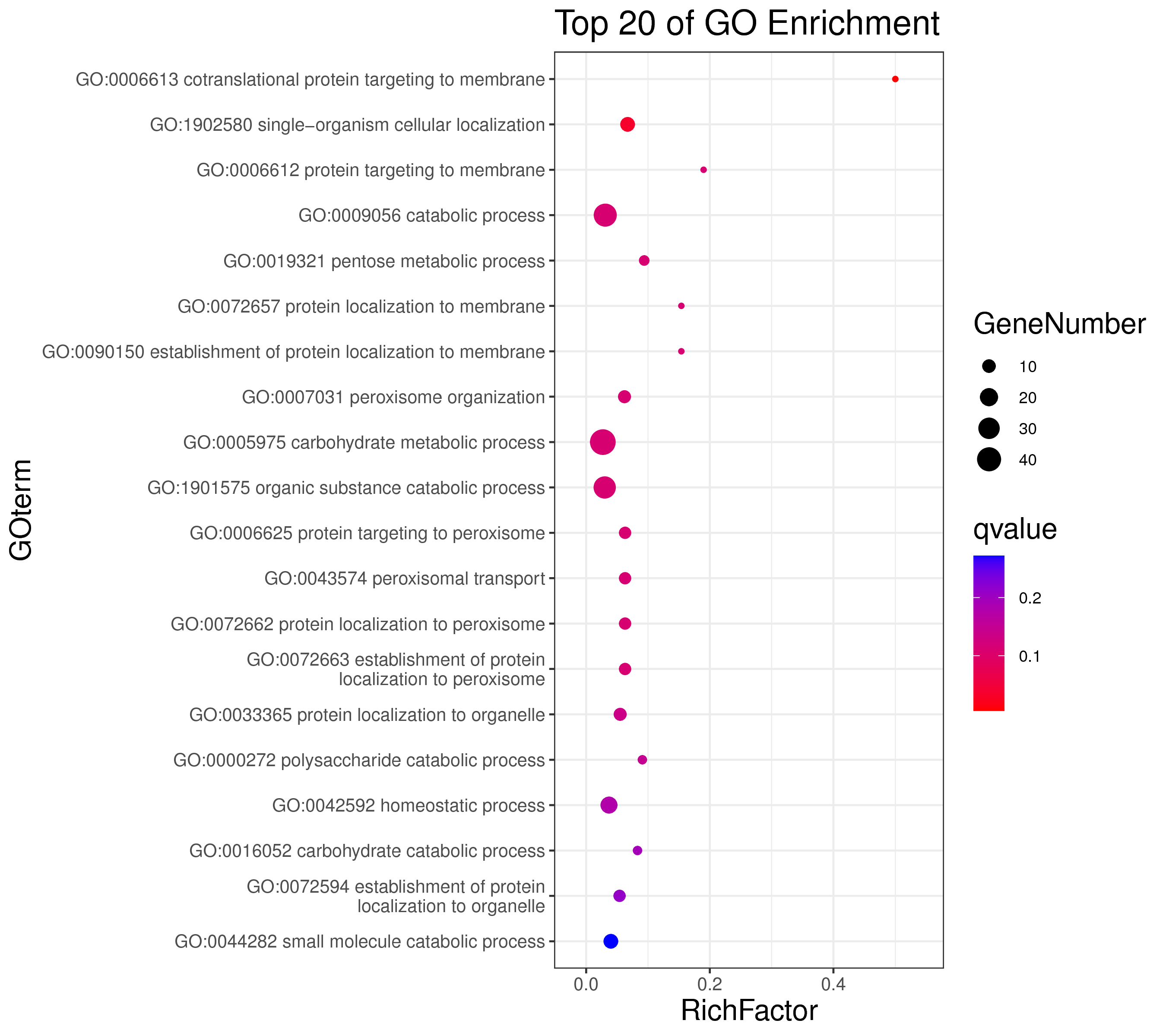
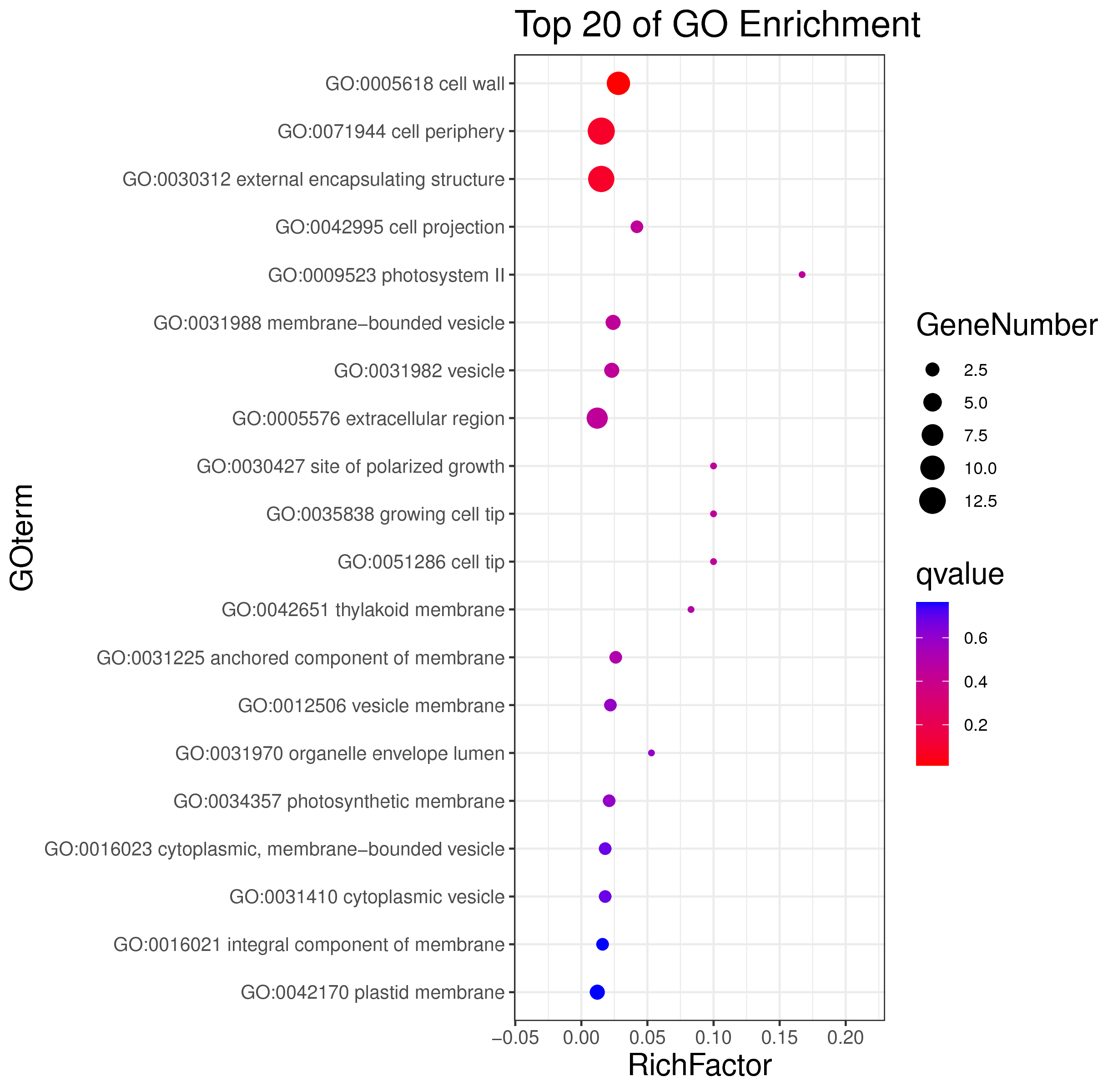
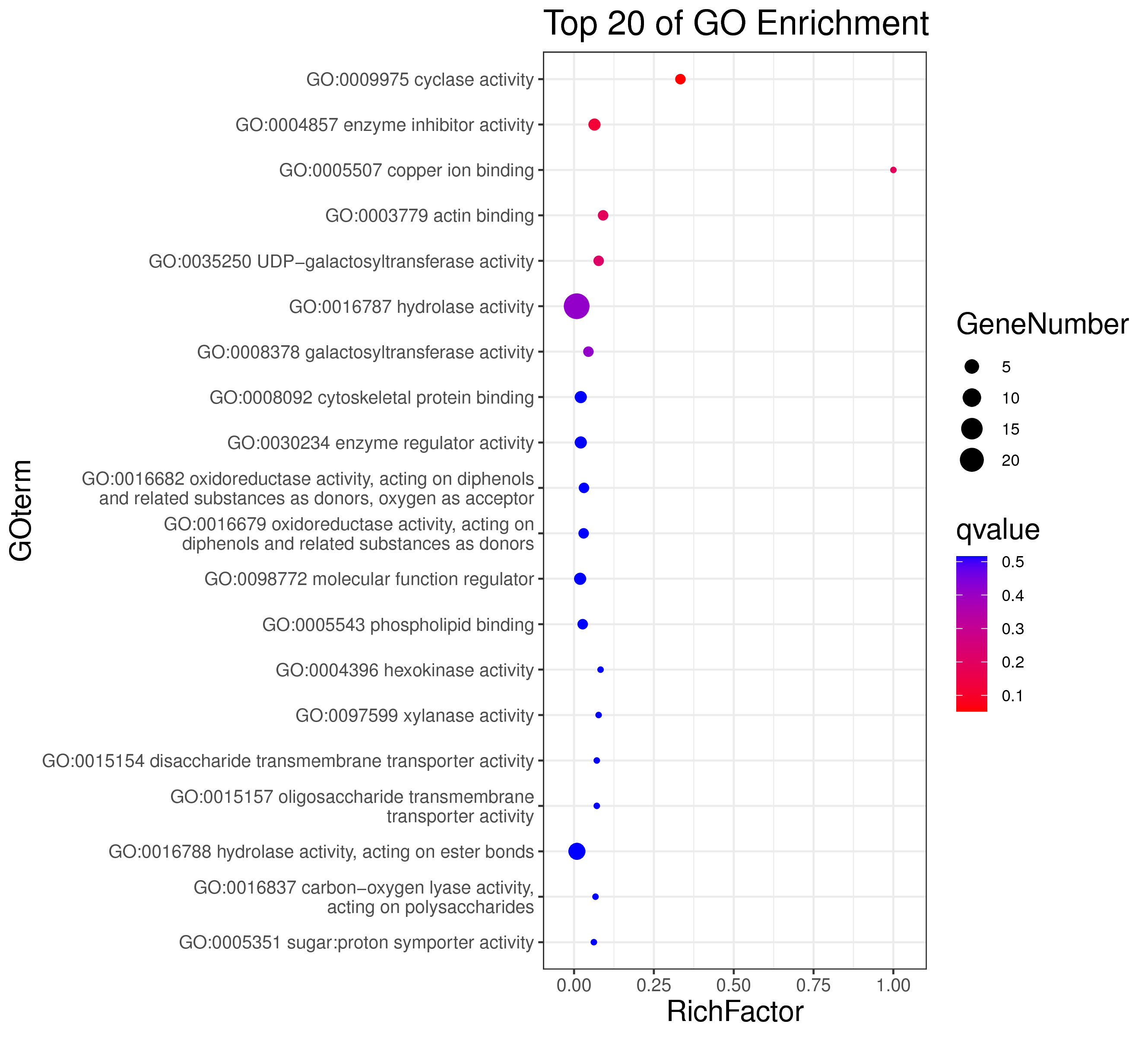
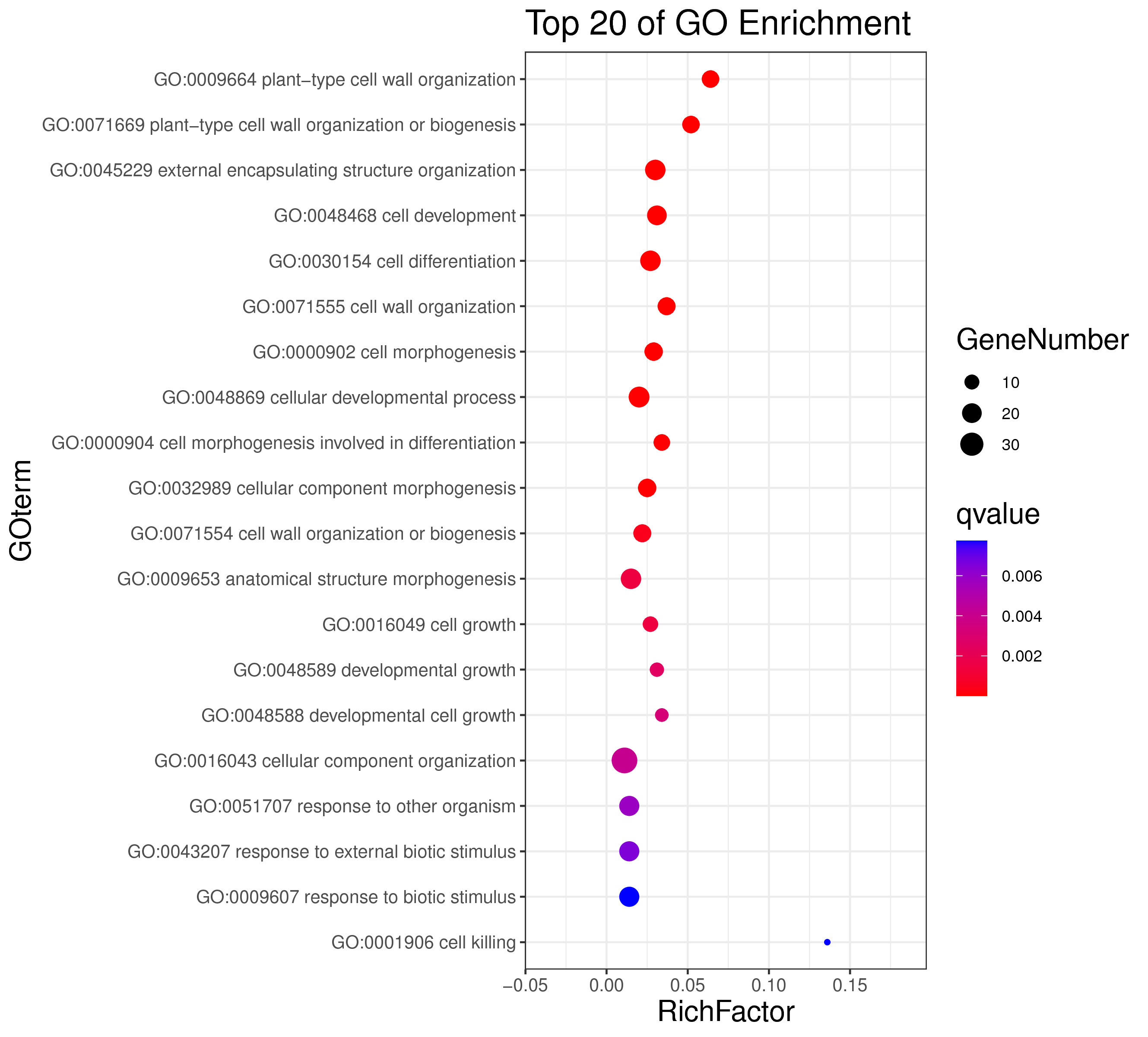
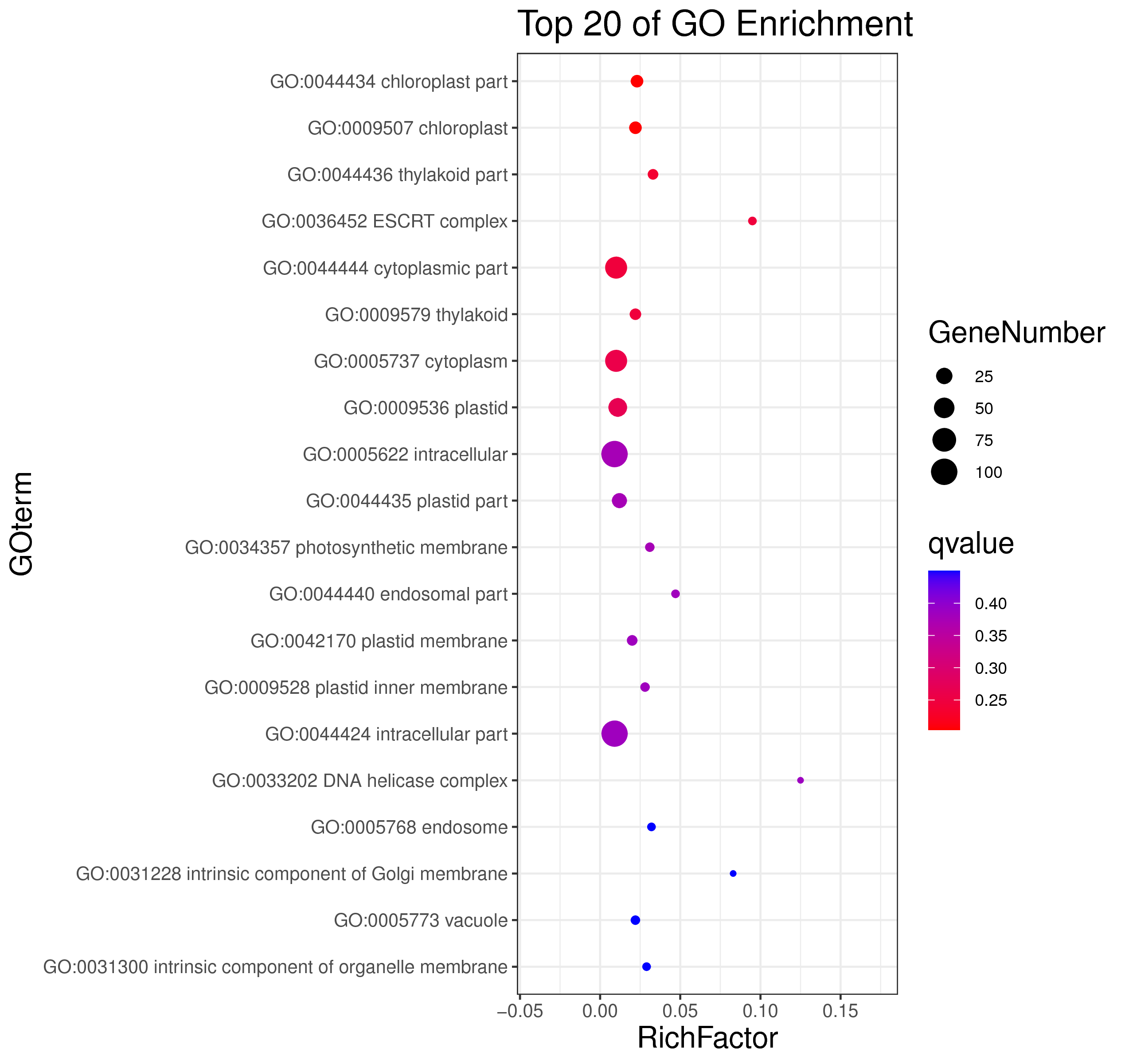
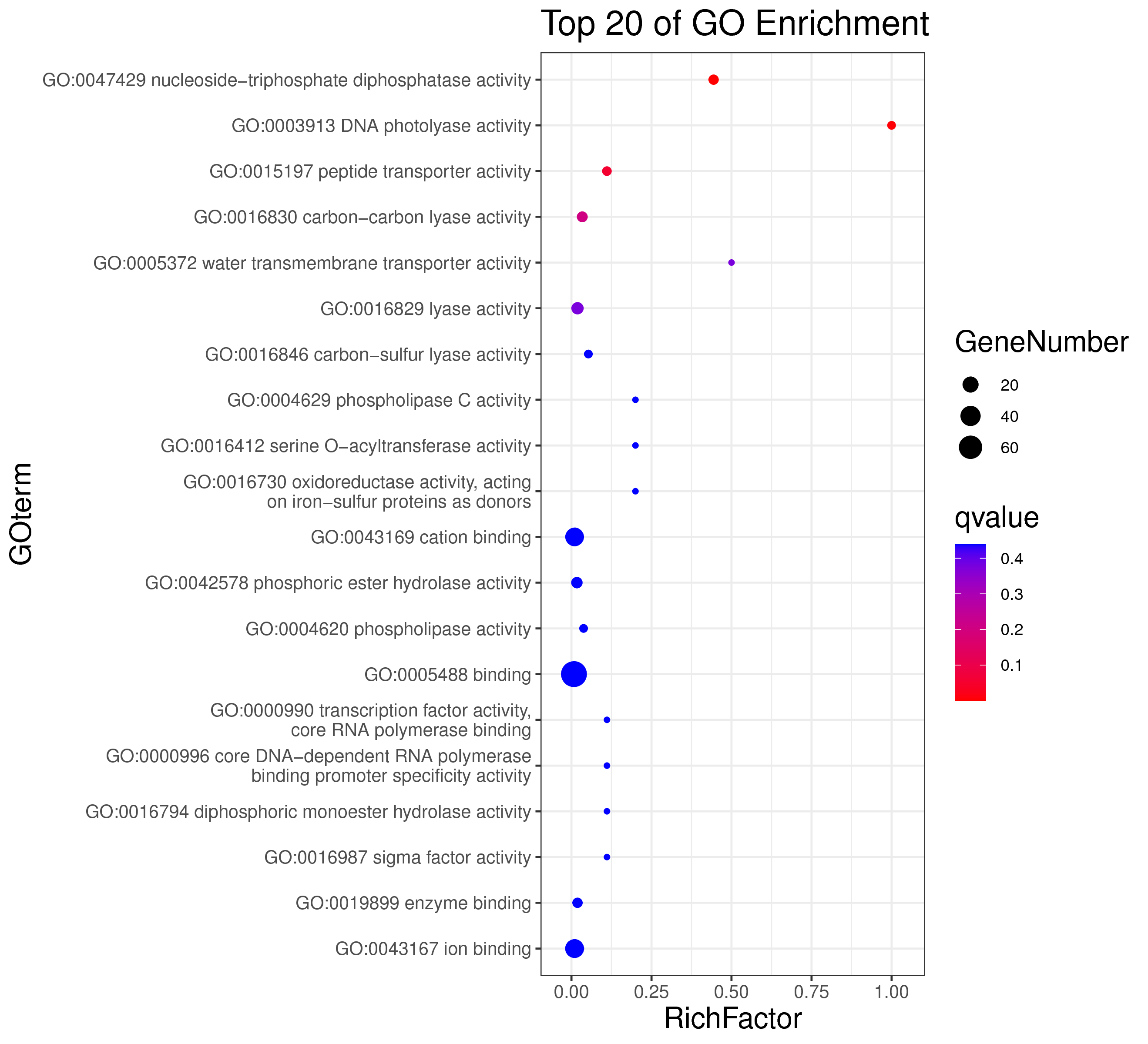
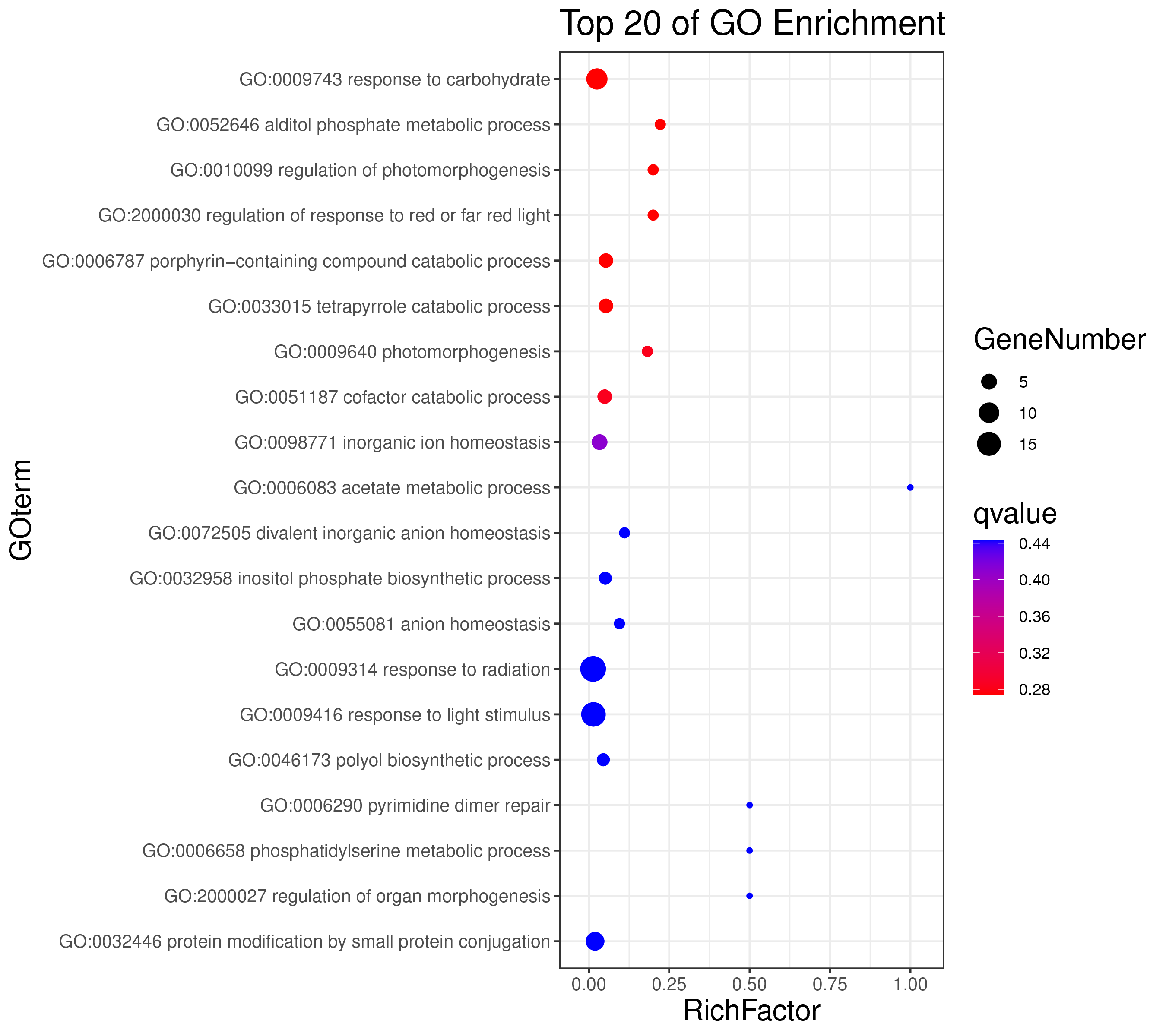
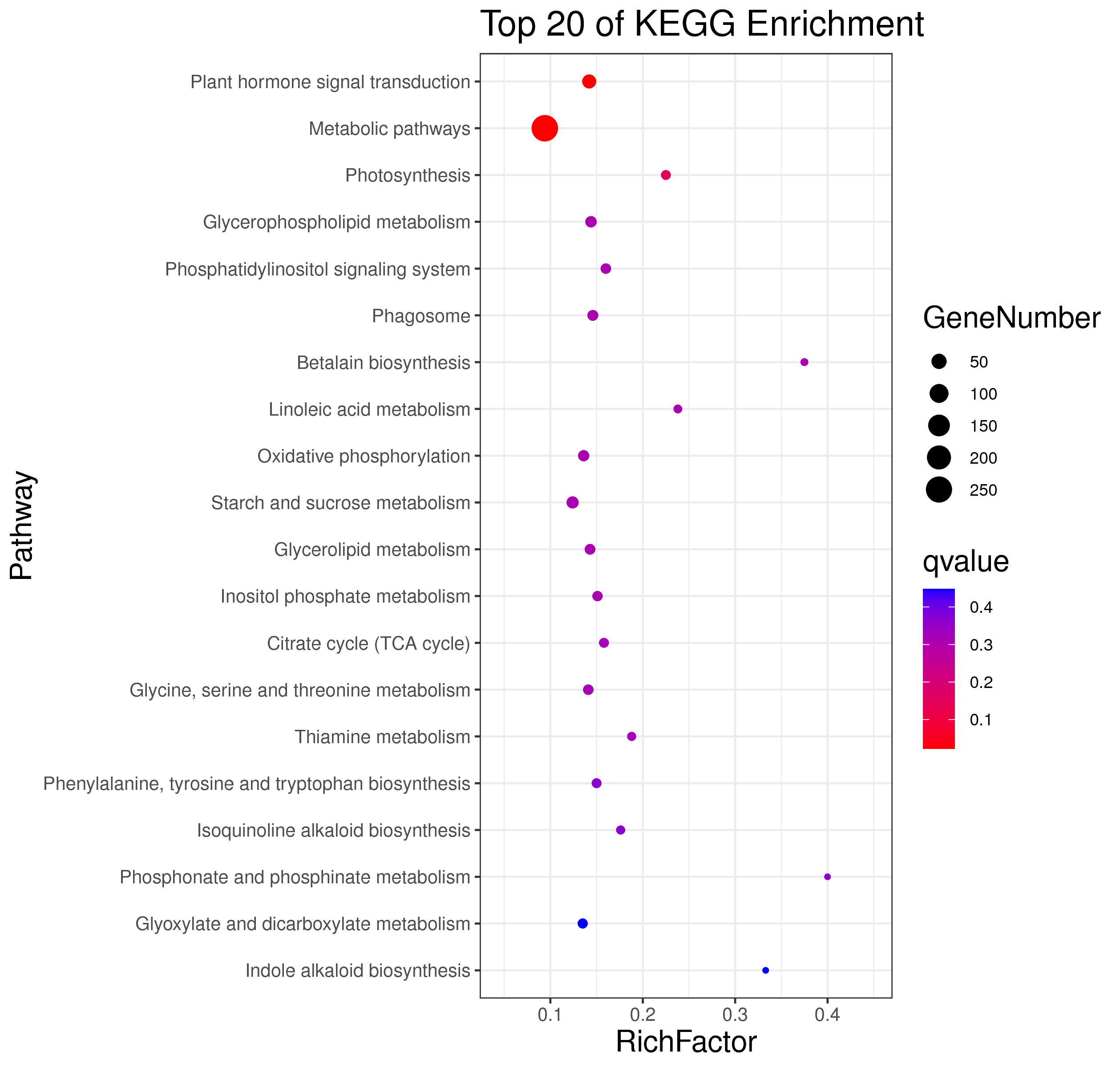
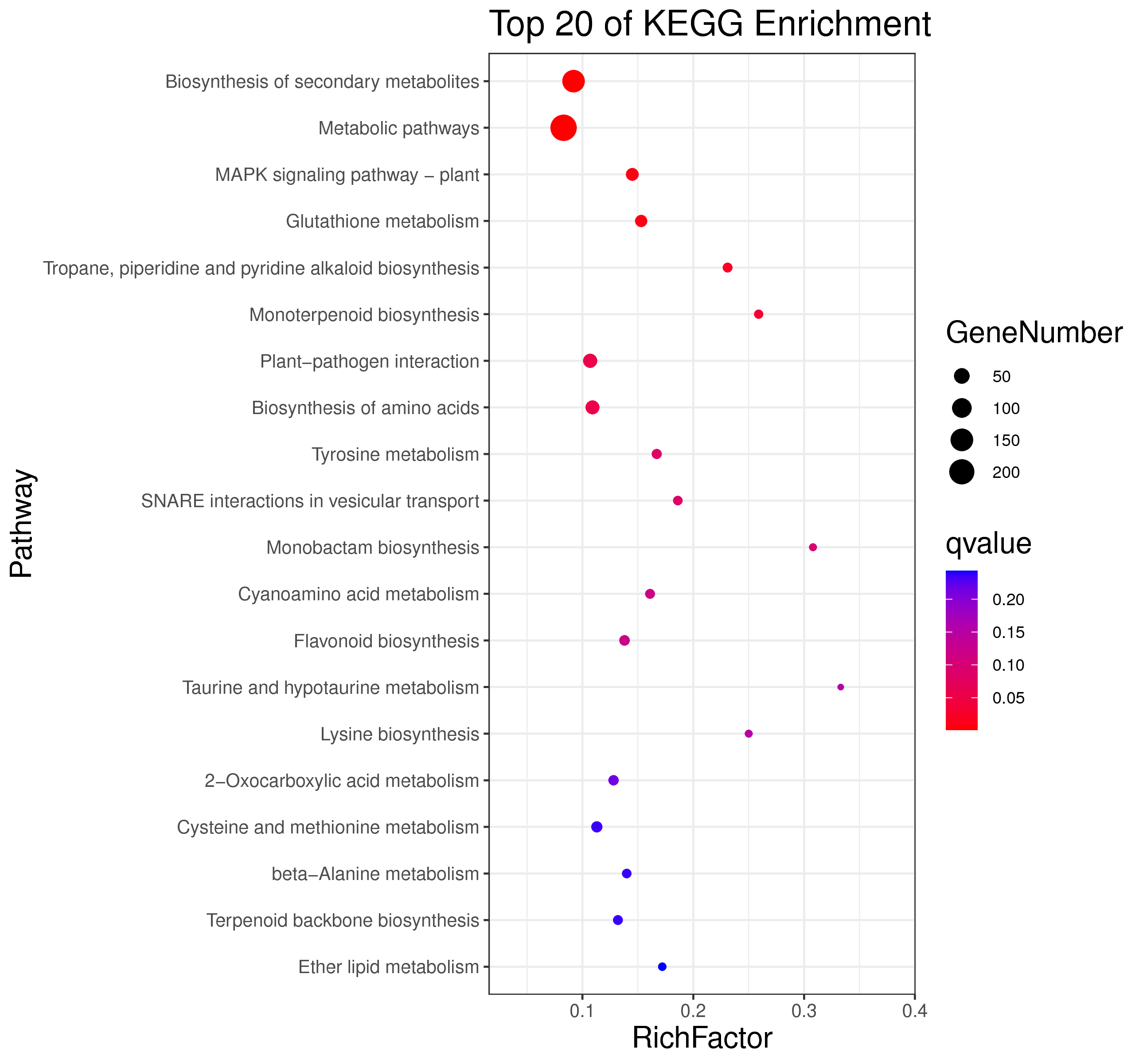
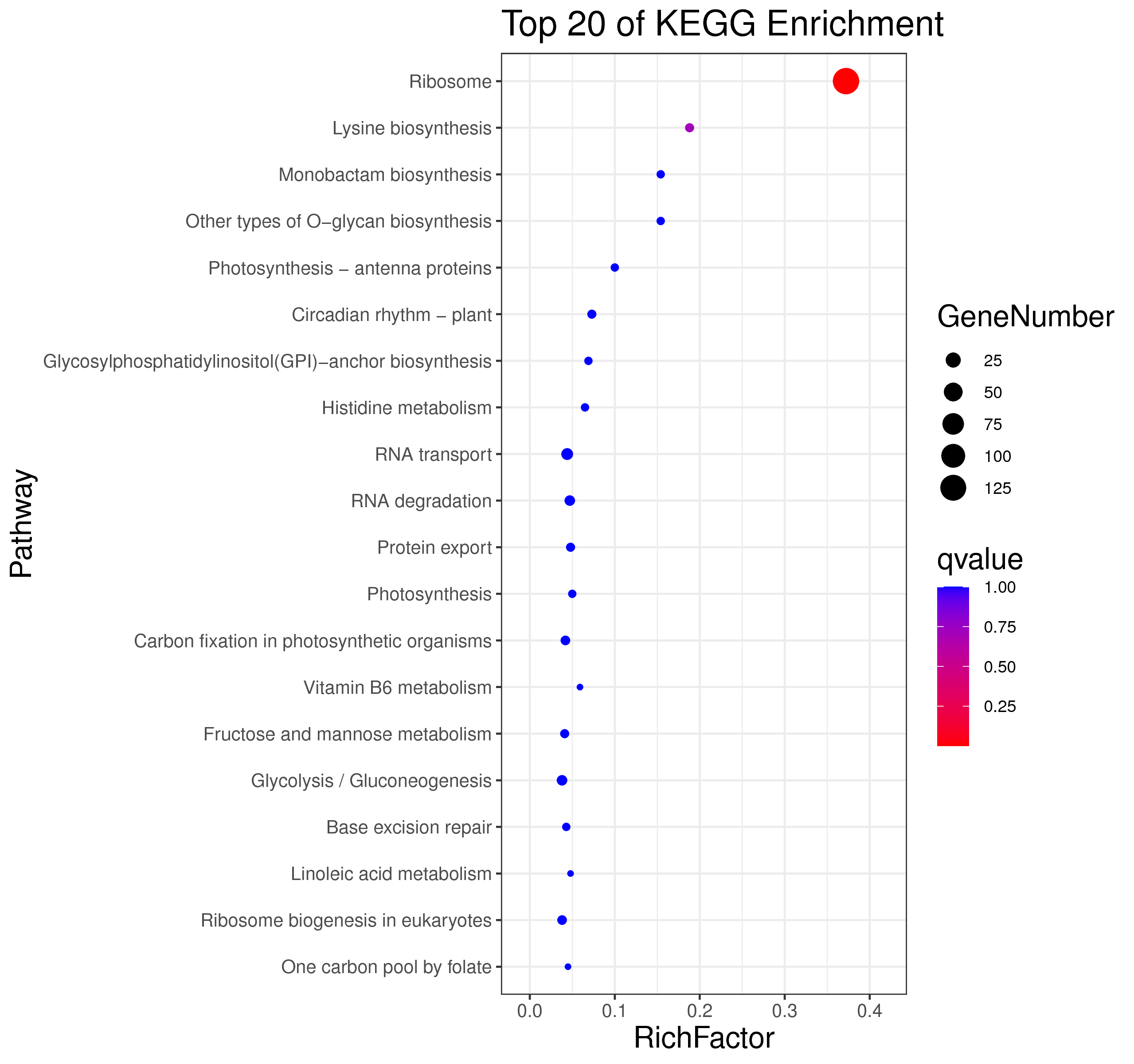
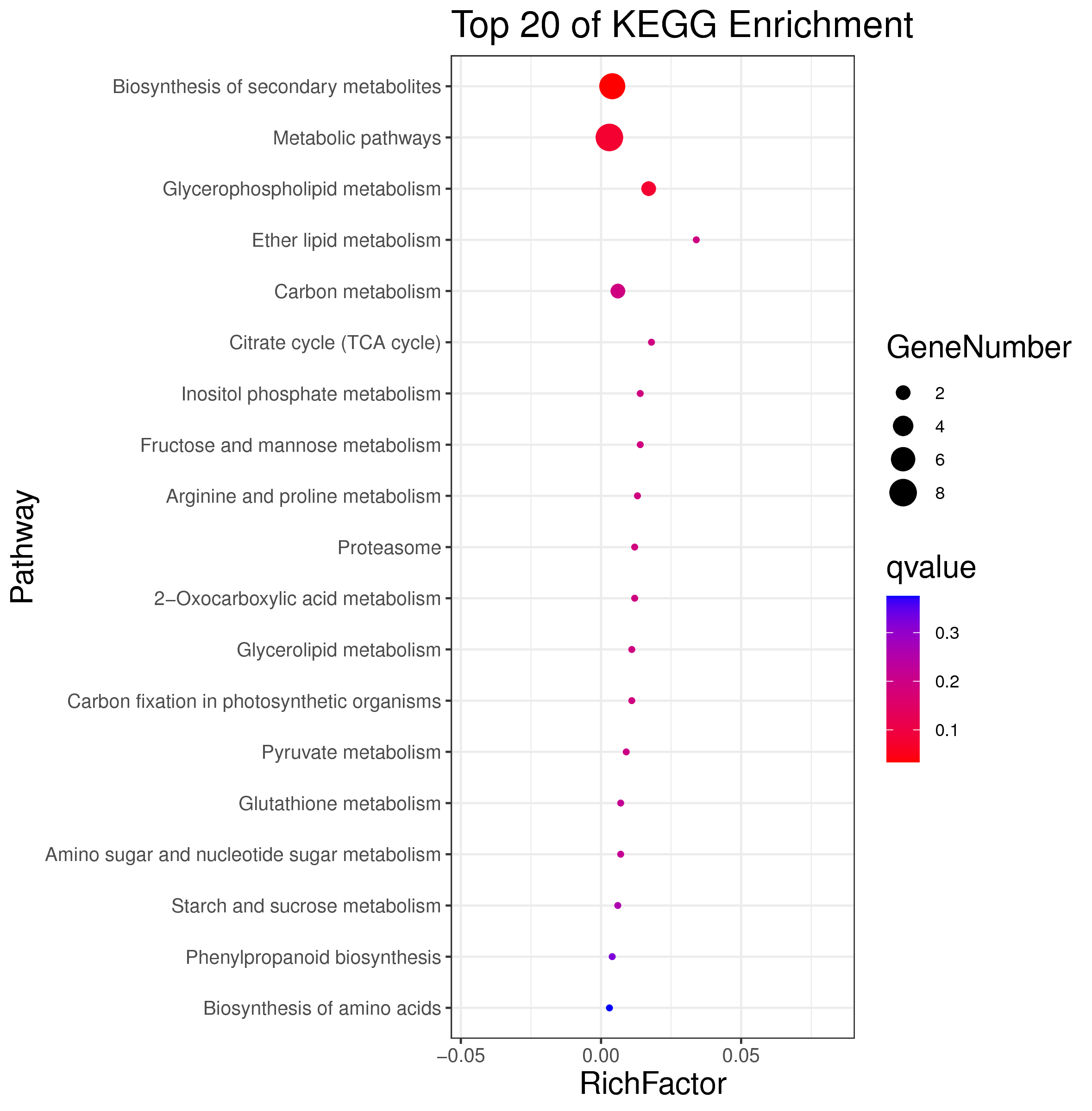
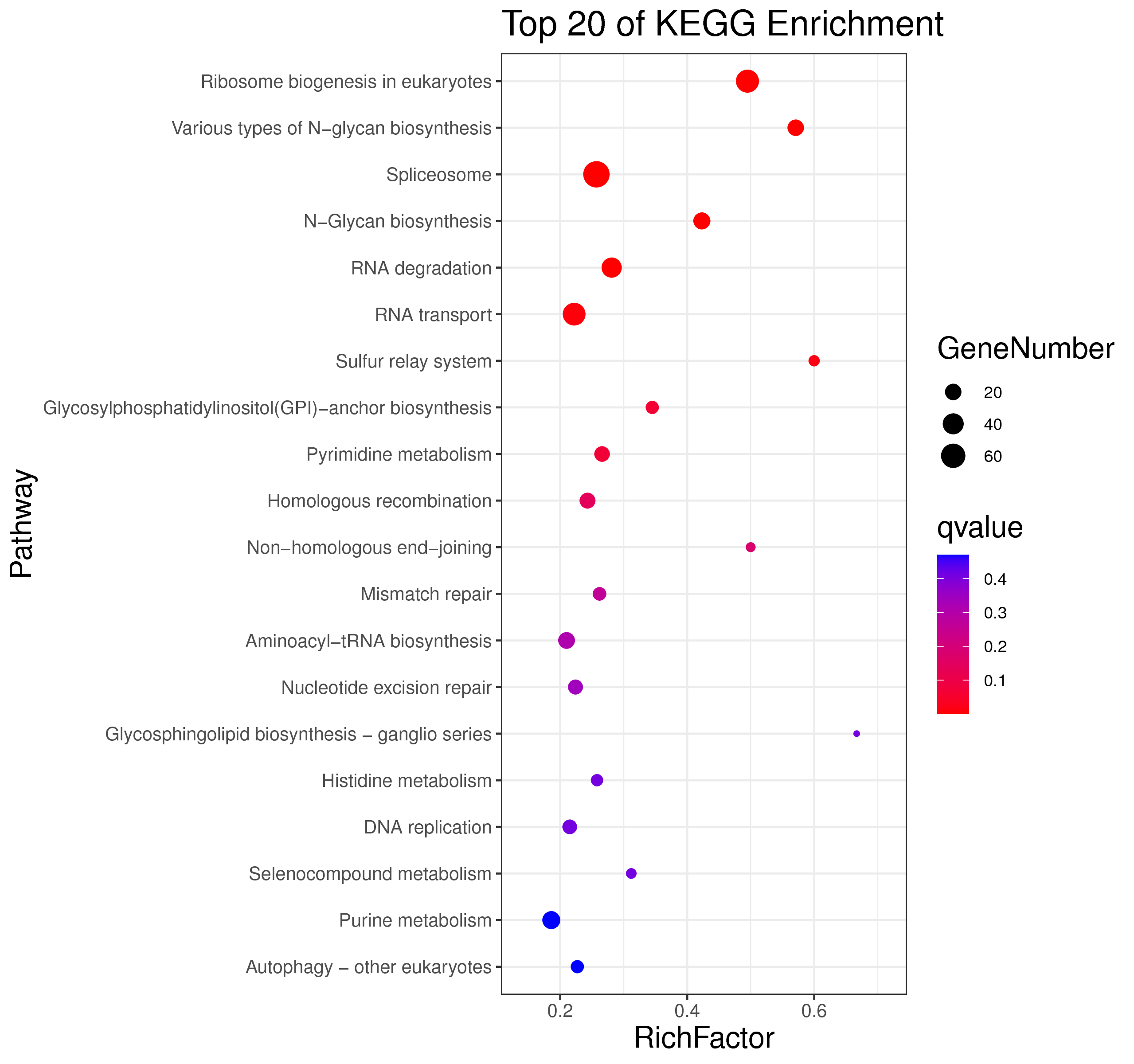
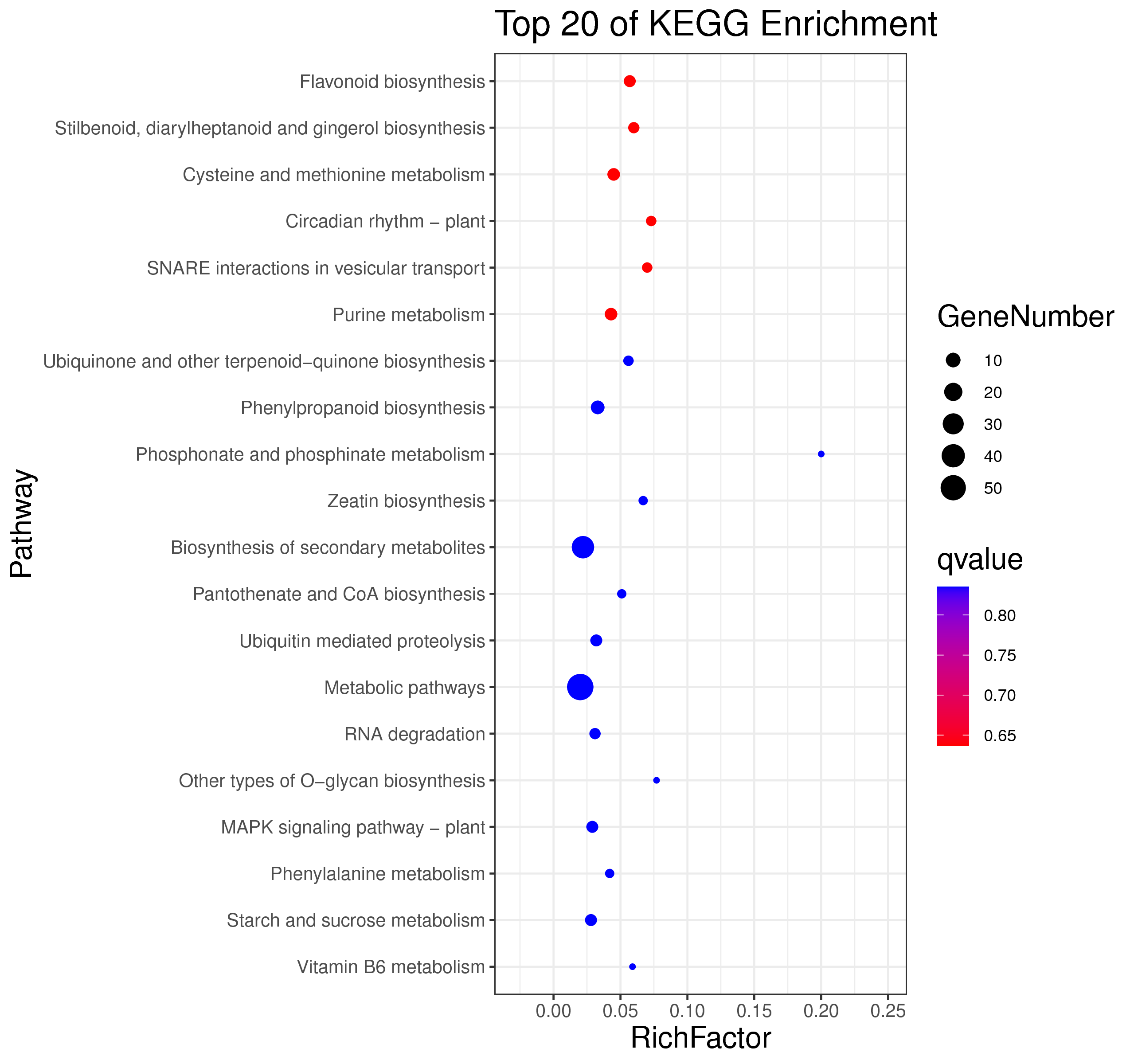
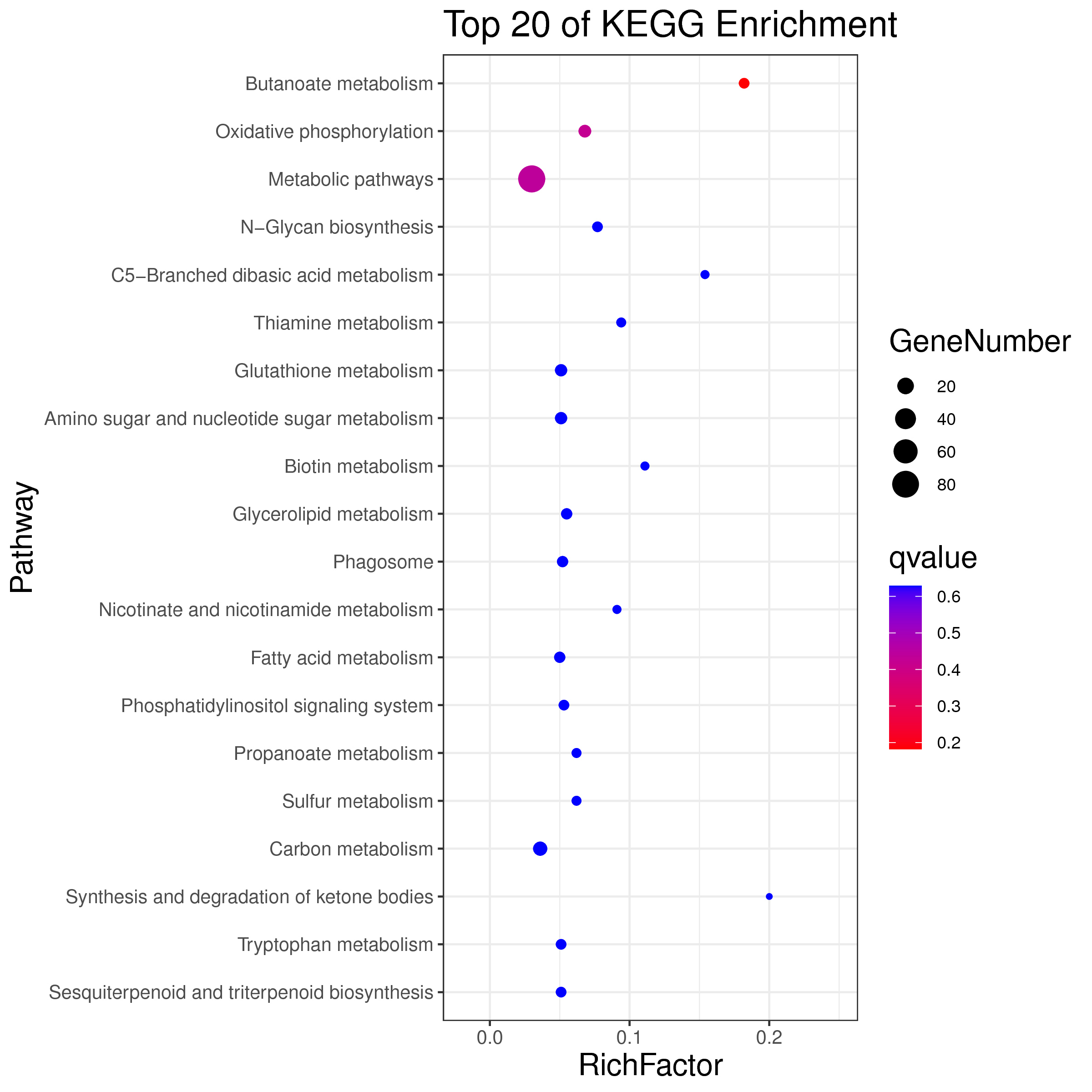
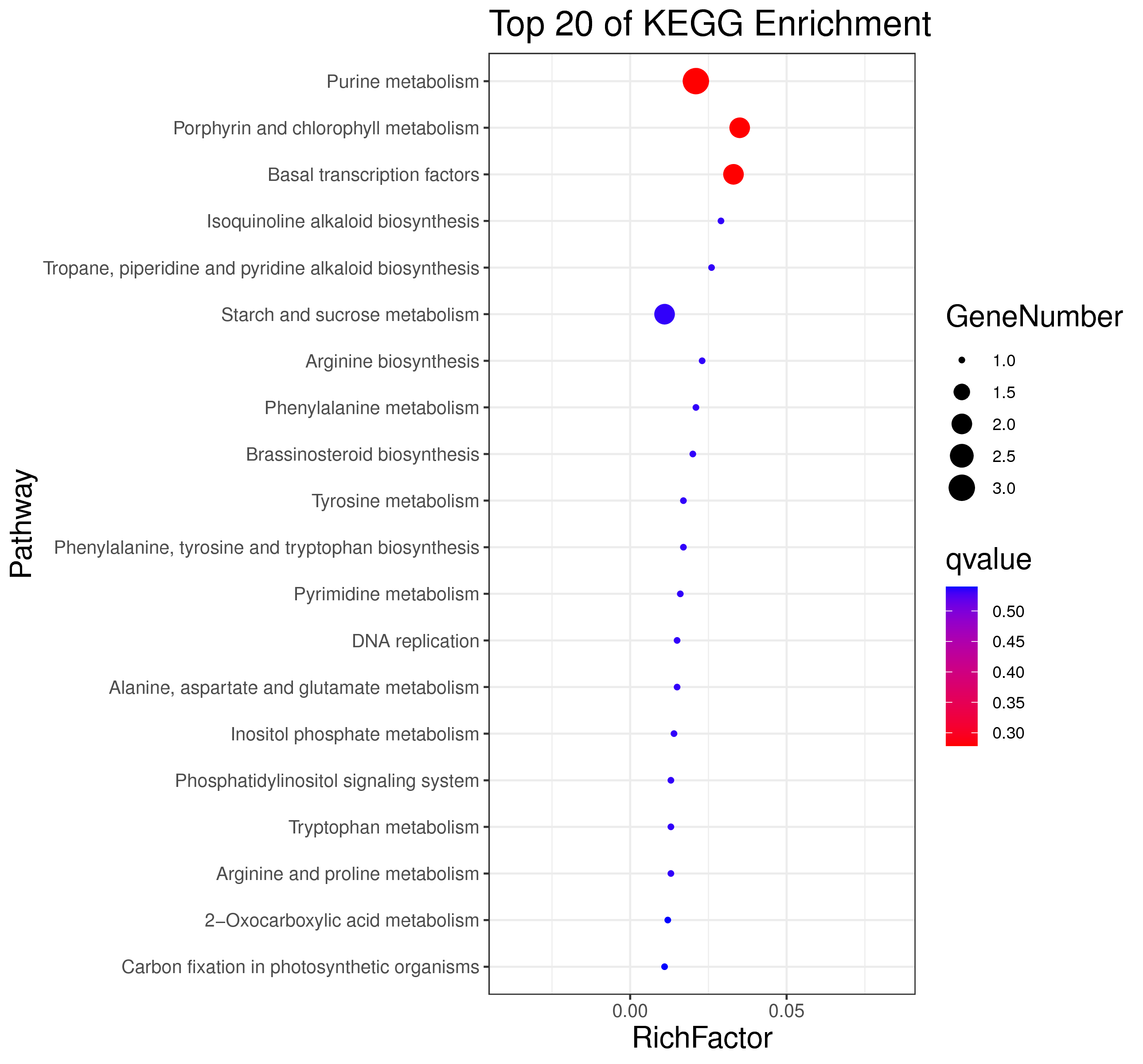
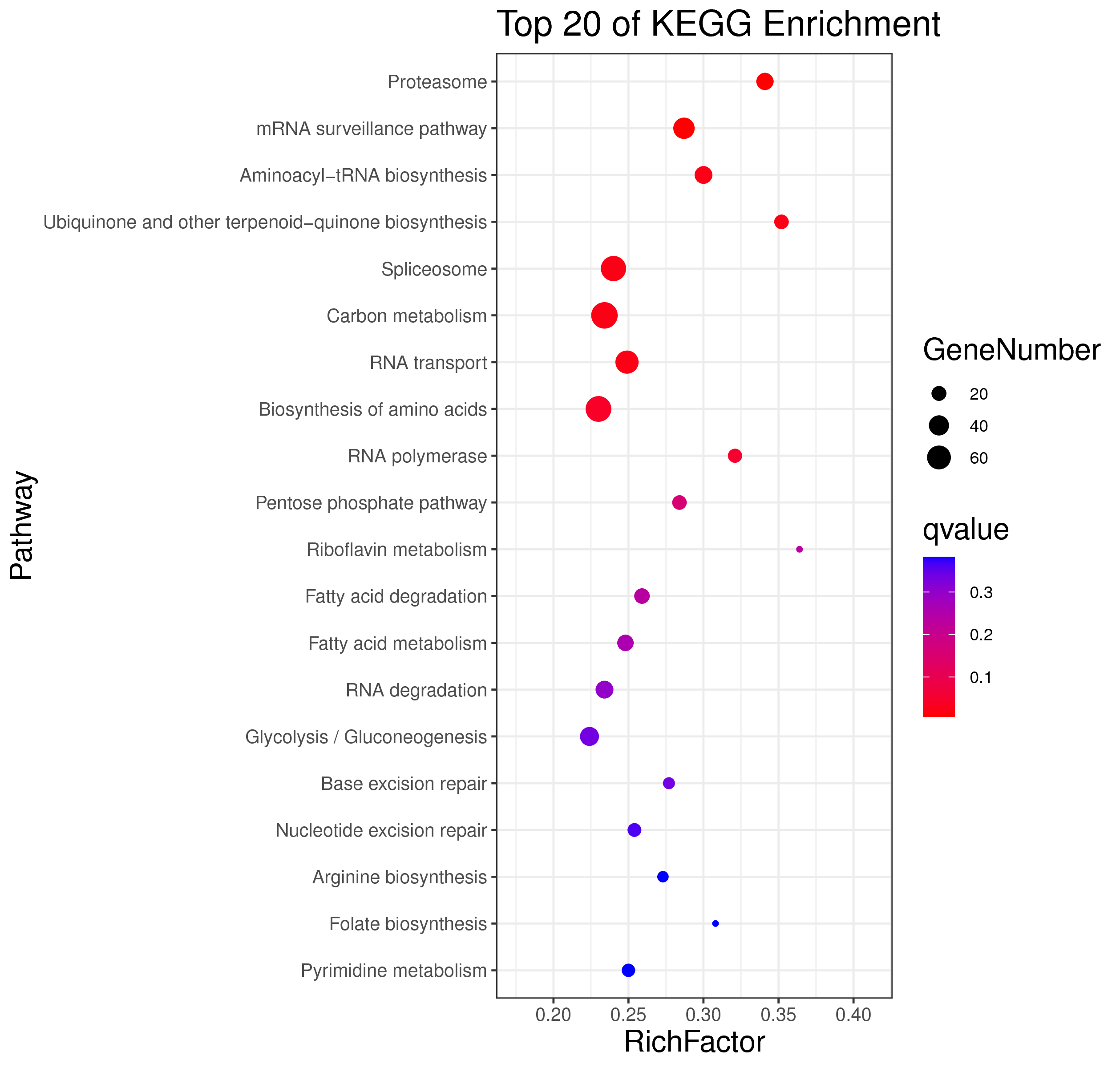
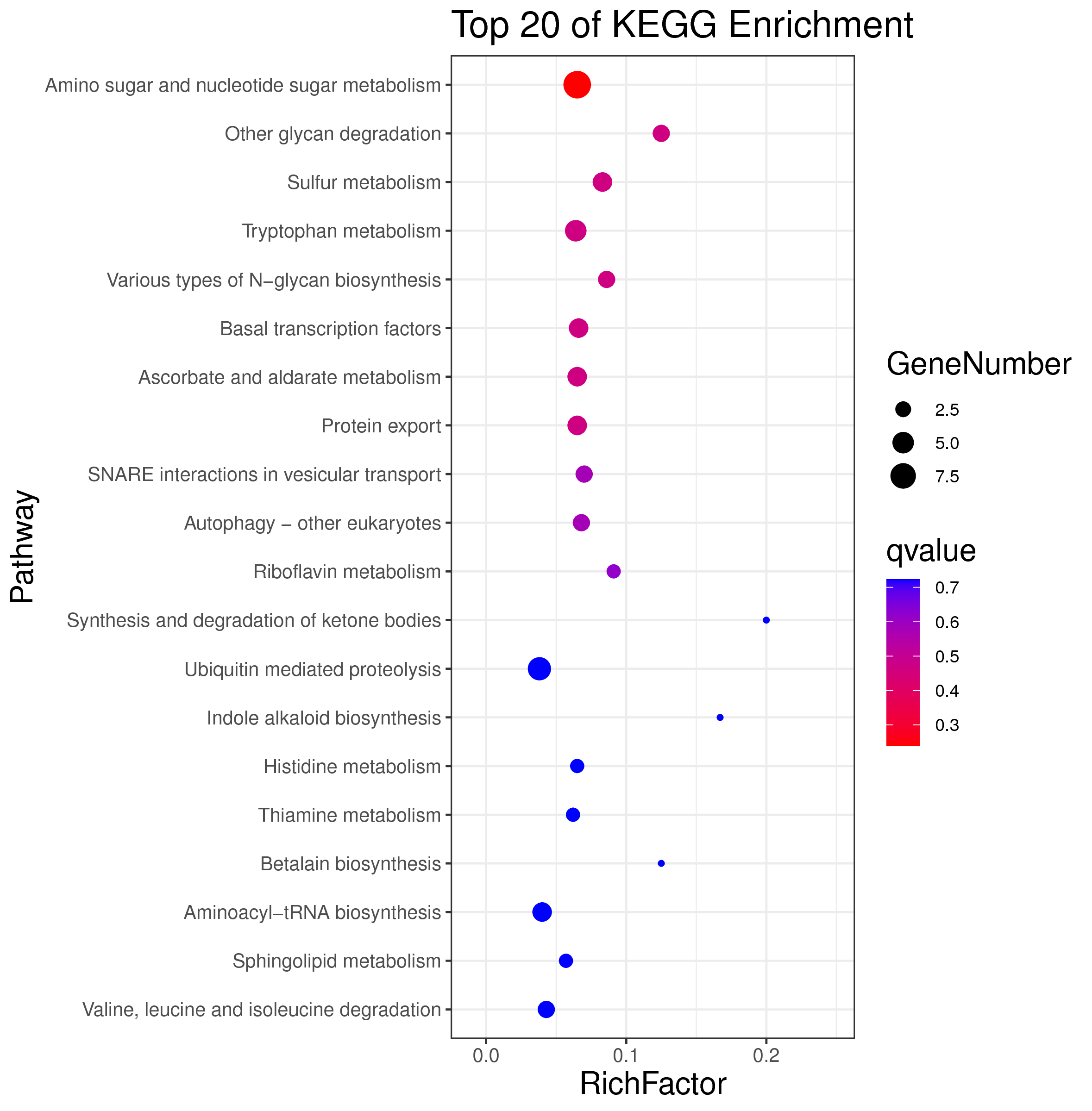
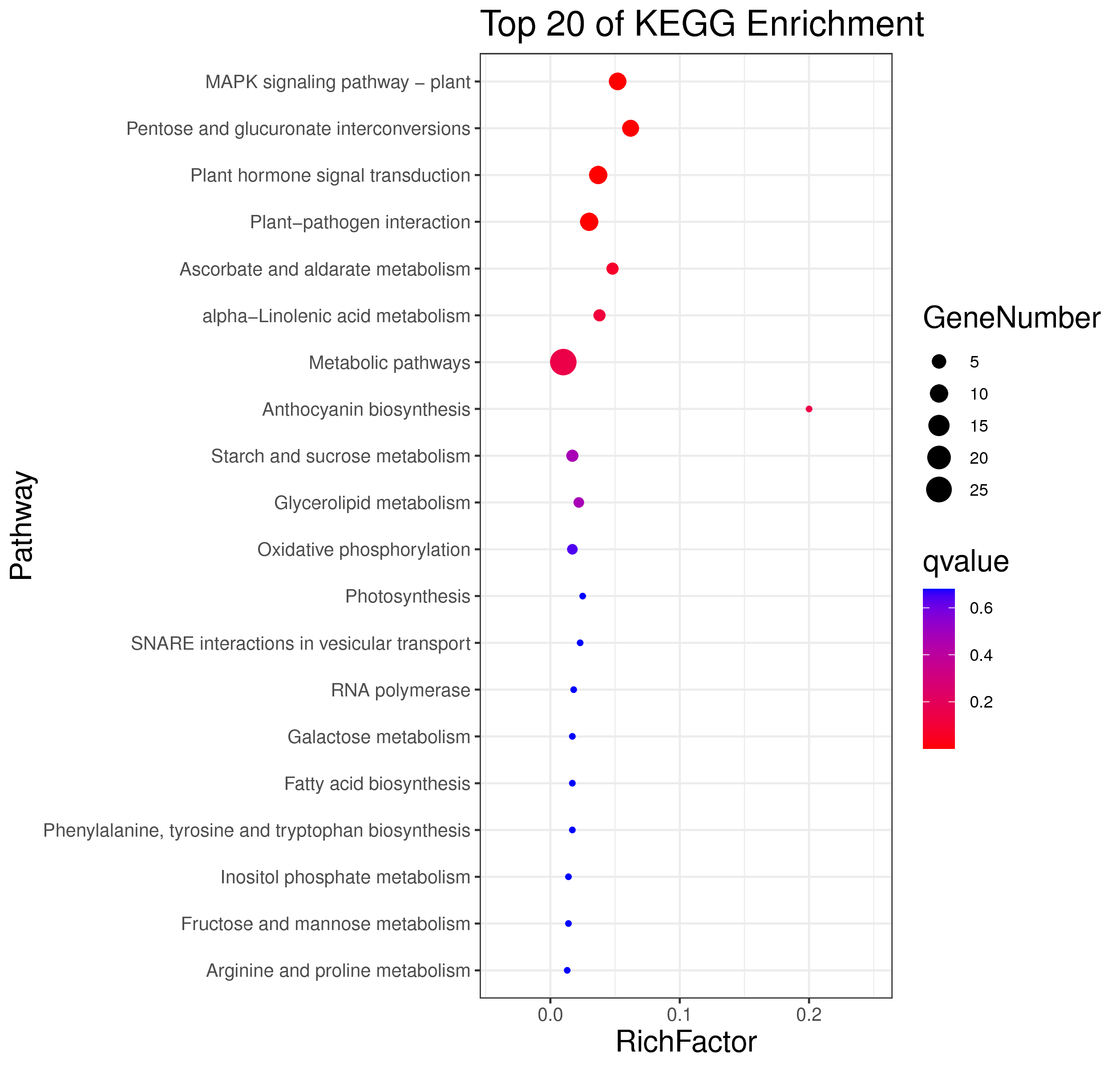
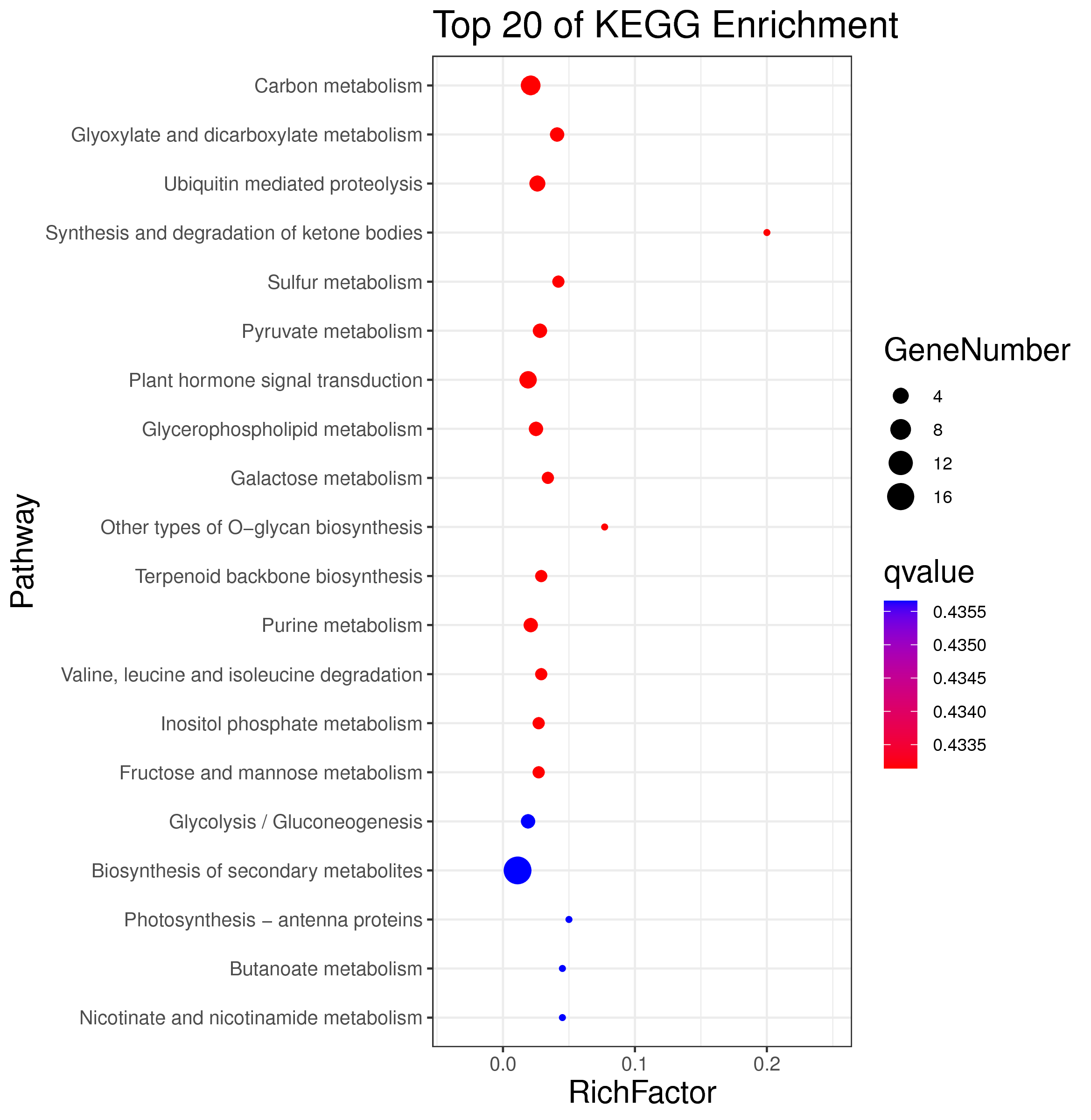
├── 5.enrichment 富集分析
│ |-- *.glist 各模块基因列表
│ |-- KO 各模块基因KO富集分析
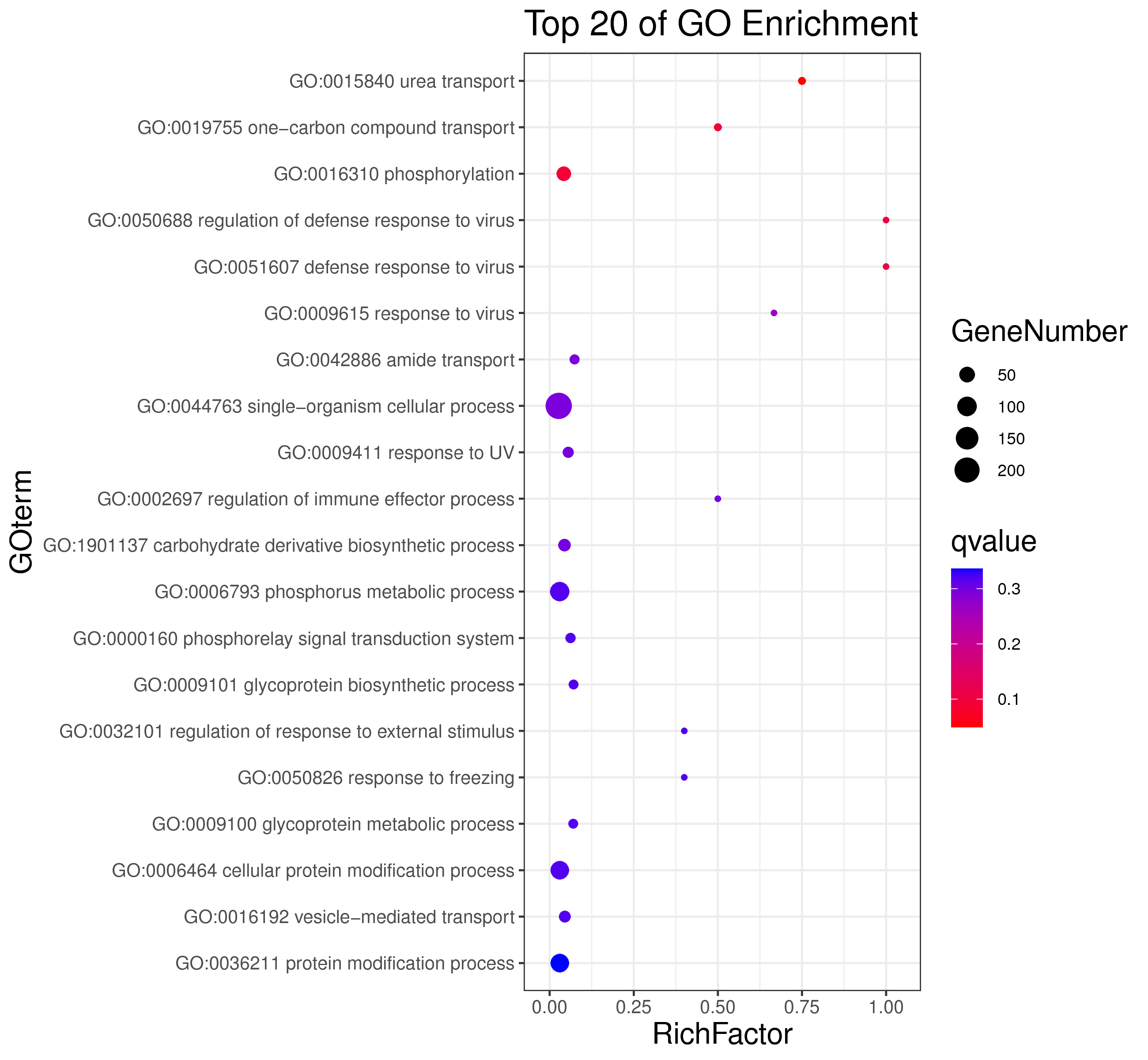
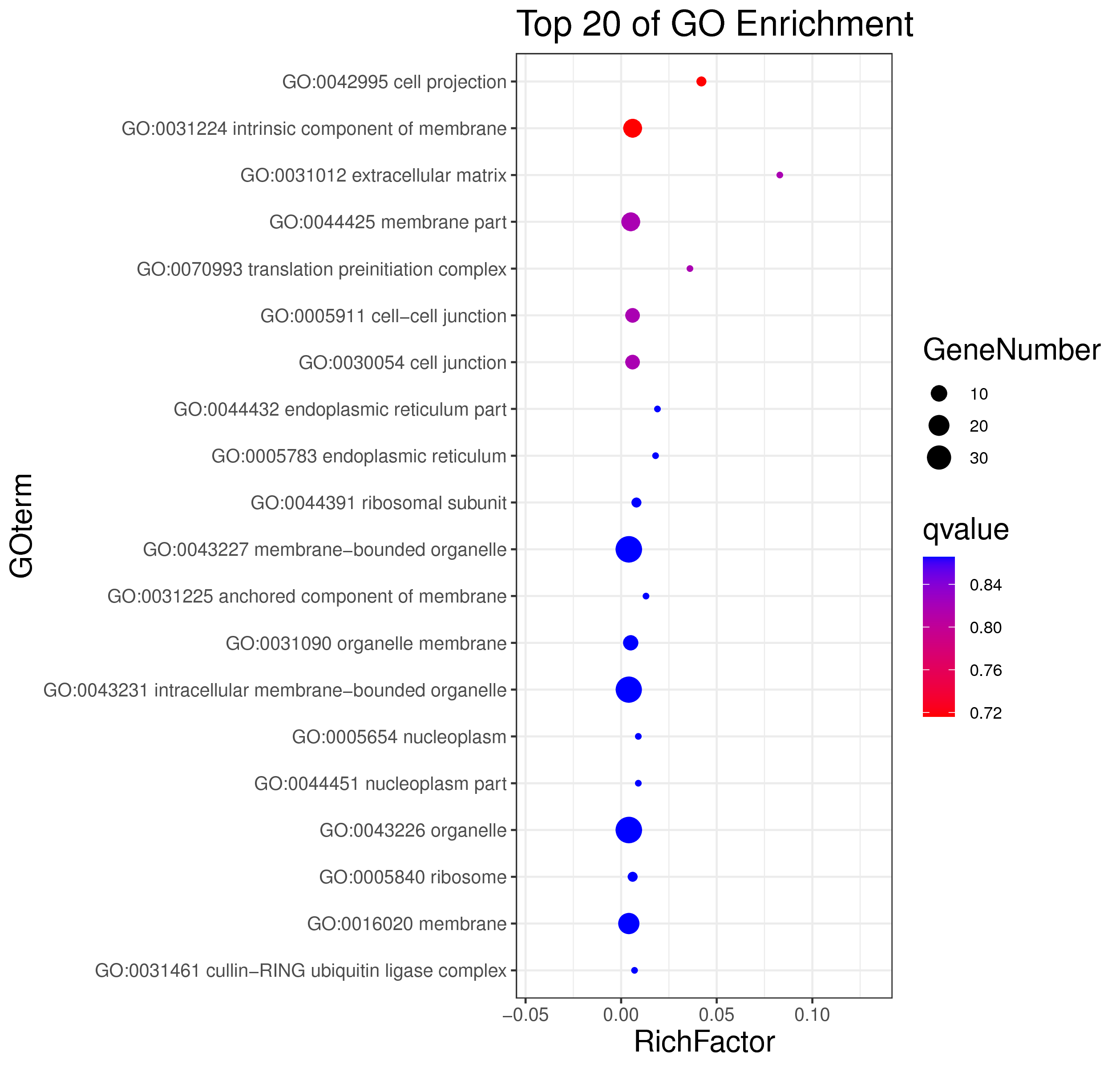
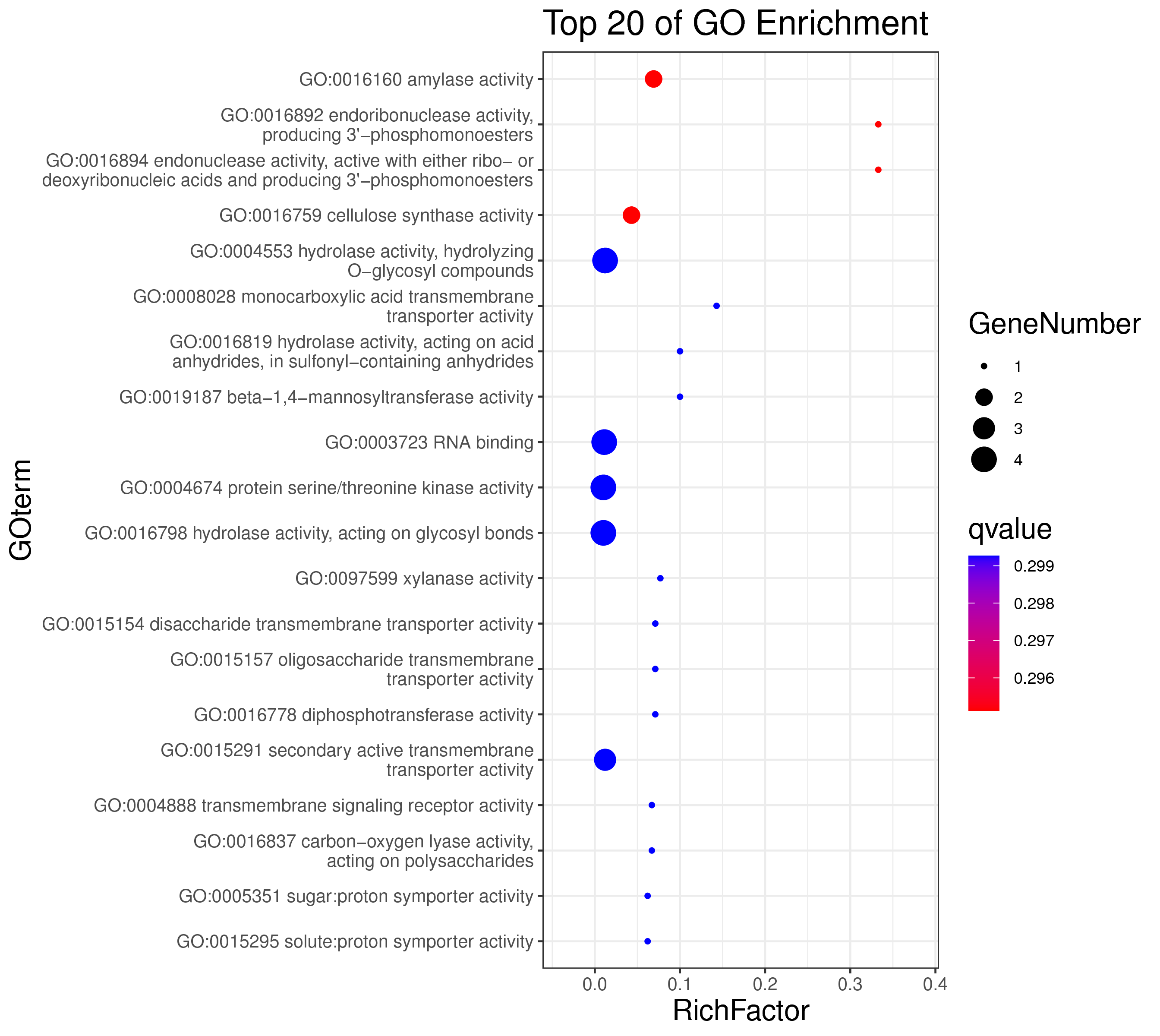
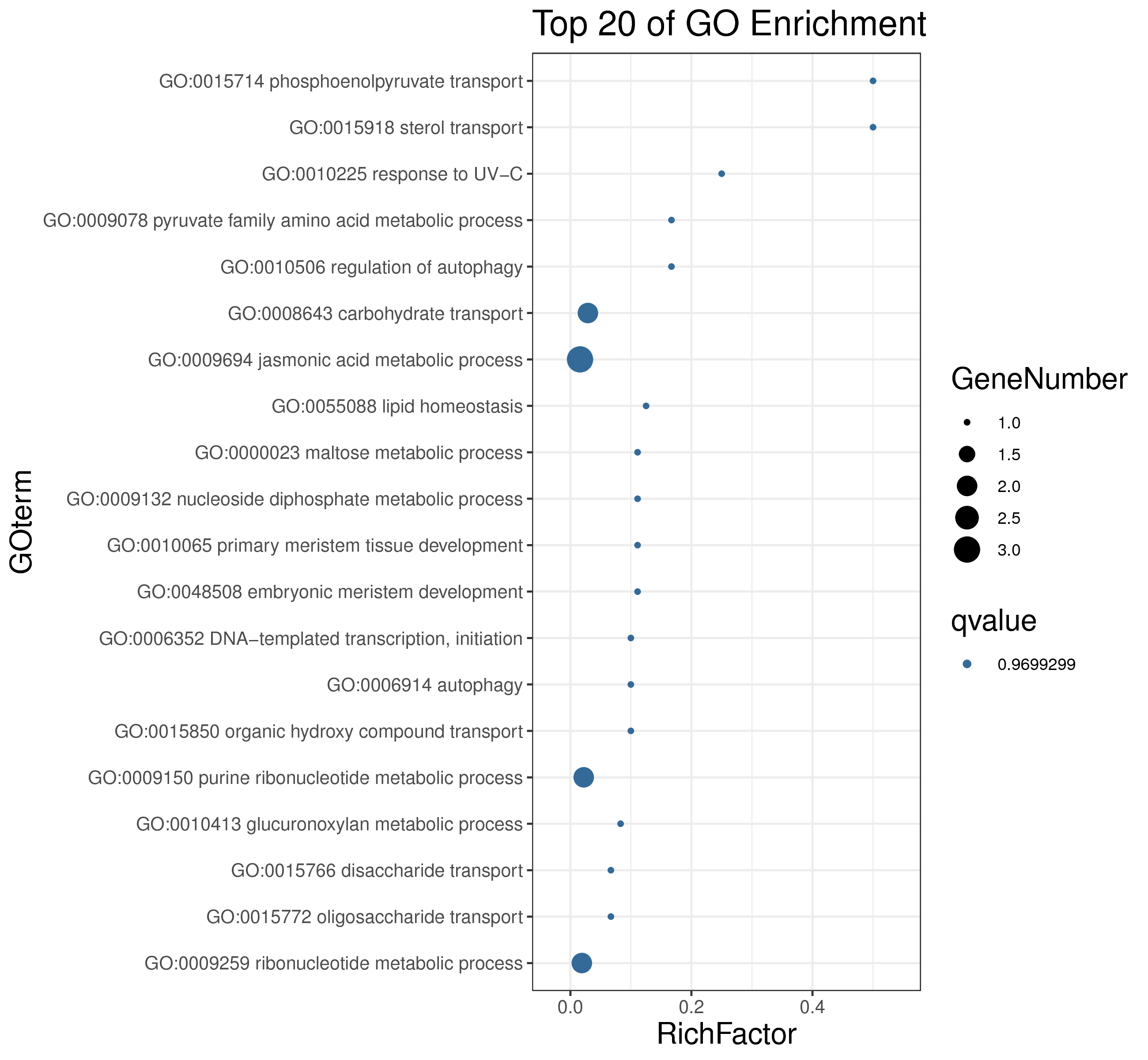
│ |-- GO 各模块基因GO富集分析
├── src 结果报告内容
│ |-- css 结题报告css脚本
│ |-- js 结题报告js脚本
│ |-- doc 结题报告说明文档
│ |-- image 结题报告图片
├── index.html 网页版结题报告
9.3 文章引用与致谢
论文引用:
如果您的研究课题使用了基迪奥的测序和分析服务,我们期望您在论文发表时,在Method部分或Acknowledgements部分引用或提及基迪奥公司。以下语句可供参考:
Method部分:
The cDNA / DNA / Small RNA libraries were sequenced on the Illumina sequencing platform by Genedenovo Biotechnology Co., Ltd (Guangzhou, China).
Acknowledgements部分:
We are grateful to/thank Guangzhou Genedenovo Biotechnology Co., Ltd for assisting in sequencing and/or bioinformatics analysis.