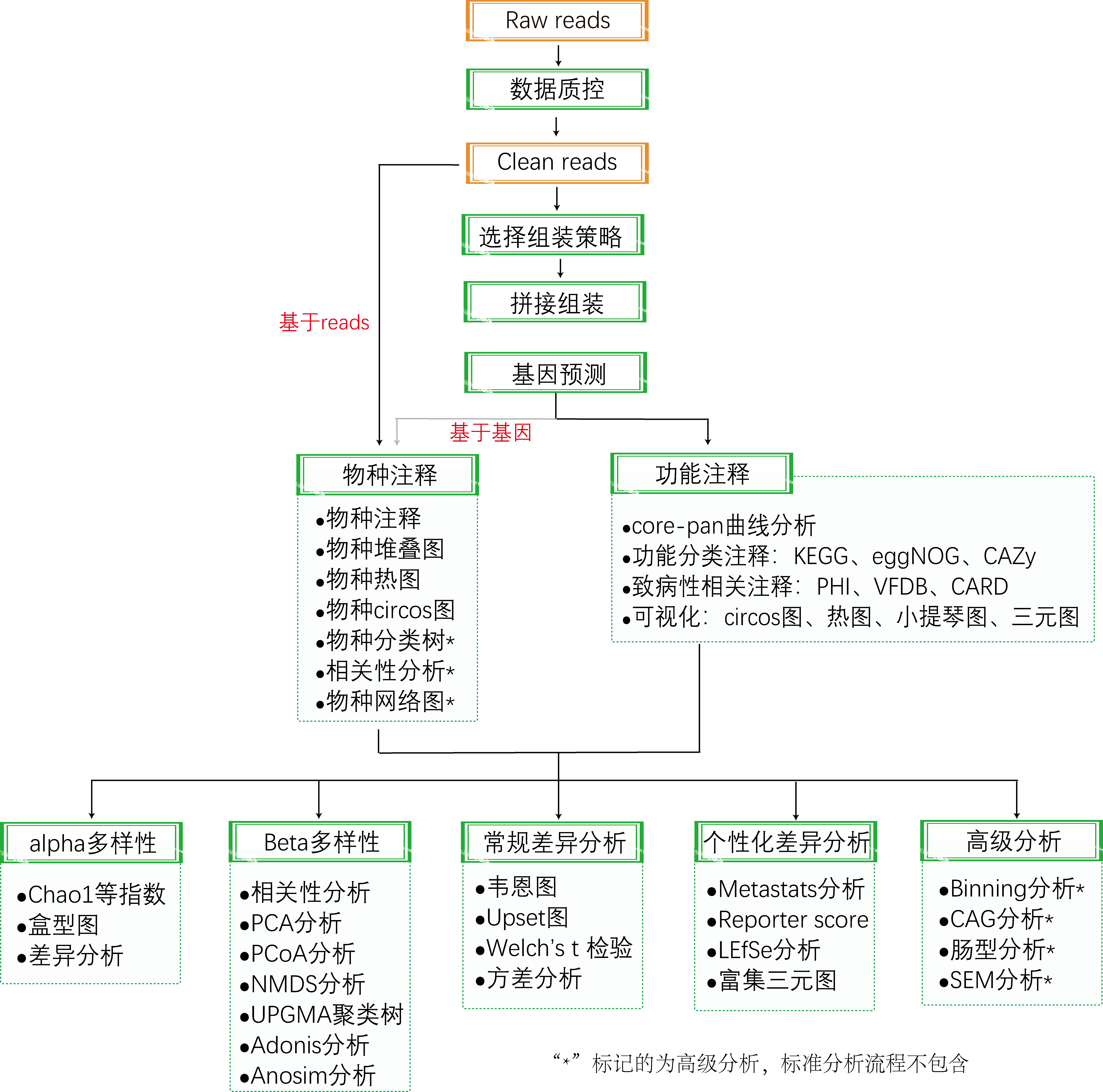
1 项目概述
| 项目编号 | GDD20090230-3_sup_2 |
| 样品信息 | F-3¦F-4¦F-5¦F-6¦G-2¦G-4¦G-5¦G-6 |
| 分组方案 | F :F-3&F-4&F-5&F-6¦G :G-2&G-4&G-5&G-6 |
| Welch's t test | F-vs-G |
| ANOVA | F-vs-G |
| Metastats | F-vs-G |
| LEfSe | F-vs-G |
| 序列组装 | 按分组, k-mer 21-141 |
| 物种注释 | 基于 reads |
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
2 技术介绍
宏基因组测序是一种利用高通量测序技术完成微生物群落所有物种基因组的检测和功能分析的方法。
宏基因组测序技术无需微生物的分离纯化培养,能够快速有效地获得整个微生物群落的基因信息,可以更加深入地对群落结构、物种分类、系统进化、基因功能及代谢网络等方面进行研究。此类方法不需要对微生物进行分离纯化培养,尤其适用于环境微生物、肠道微生物等样本的研究。
2.1 实验流程
![]() |
| Fig 2-1-1 实验流程图 |
2.2 分析流程
![]() |
| Fig 2-2-1 信息分析流程图 |
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
3 数据处理
3.1 数据过滤
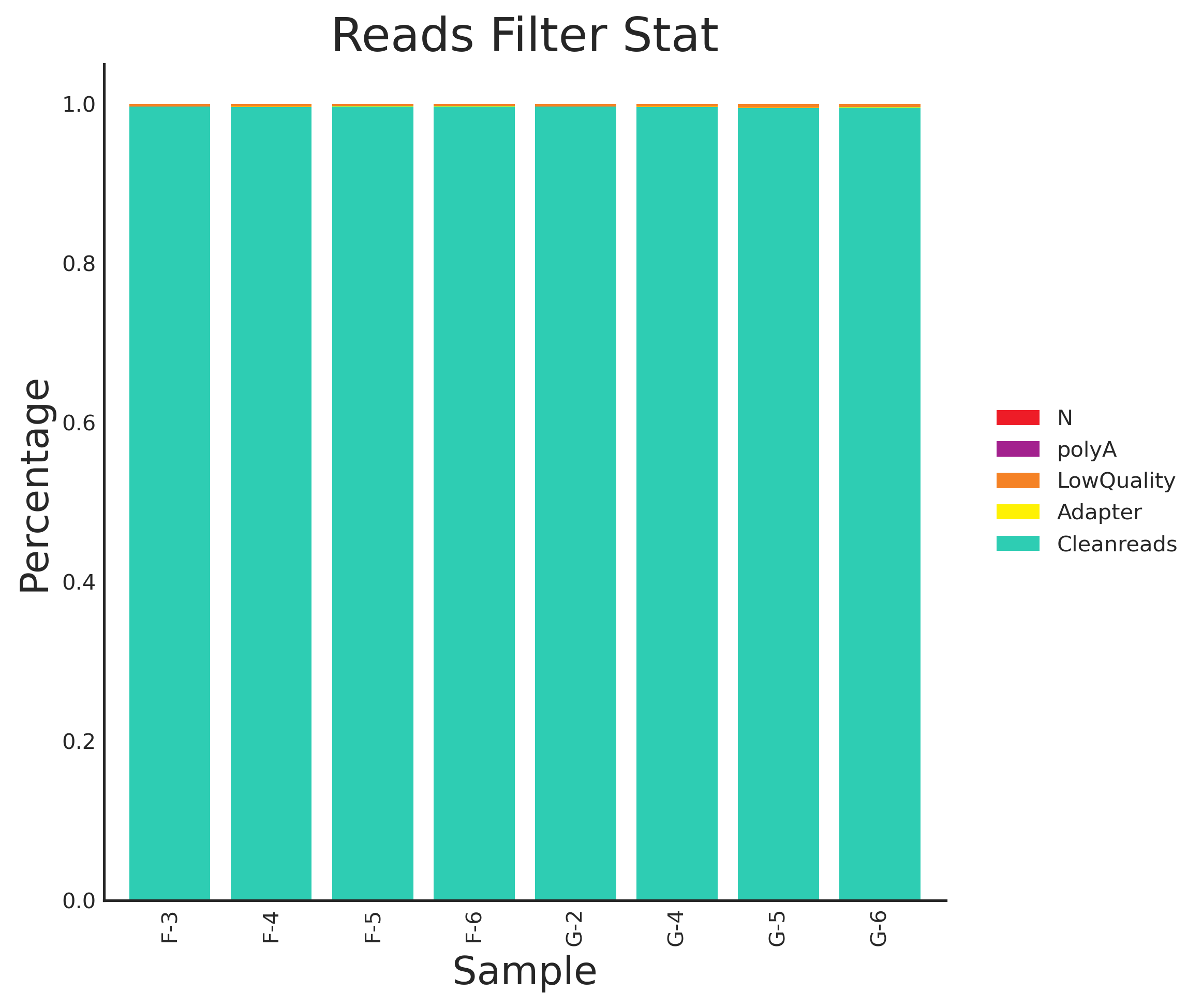
测序完成之后获得原始数据(raw data)在正常情况下会存在一部分低质量数据,这些低质量数据会影响后续分析结果的准确性。因此我们会根据一定的标准过滤掉低质量数据,从而获得用于后续准确分析的高质量数据(clean data)。
数据过滤的标准如下:
- 去除含adapter的reads;
- 去除N的比例大于10%的reads;
- 去除低质量reads(质量值Q≤20的碱基数占整个read的50%以上);
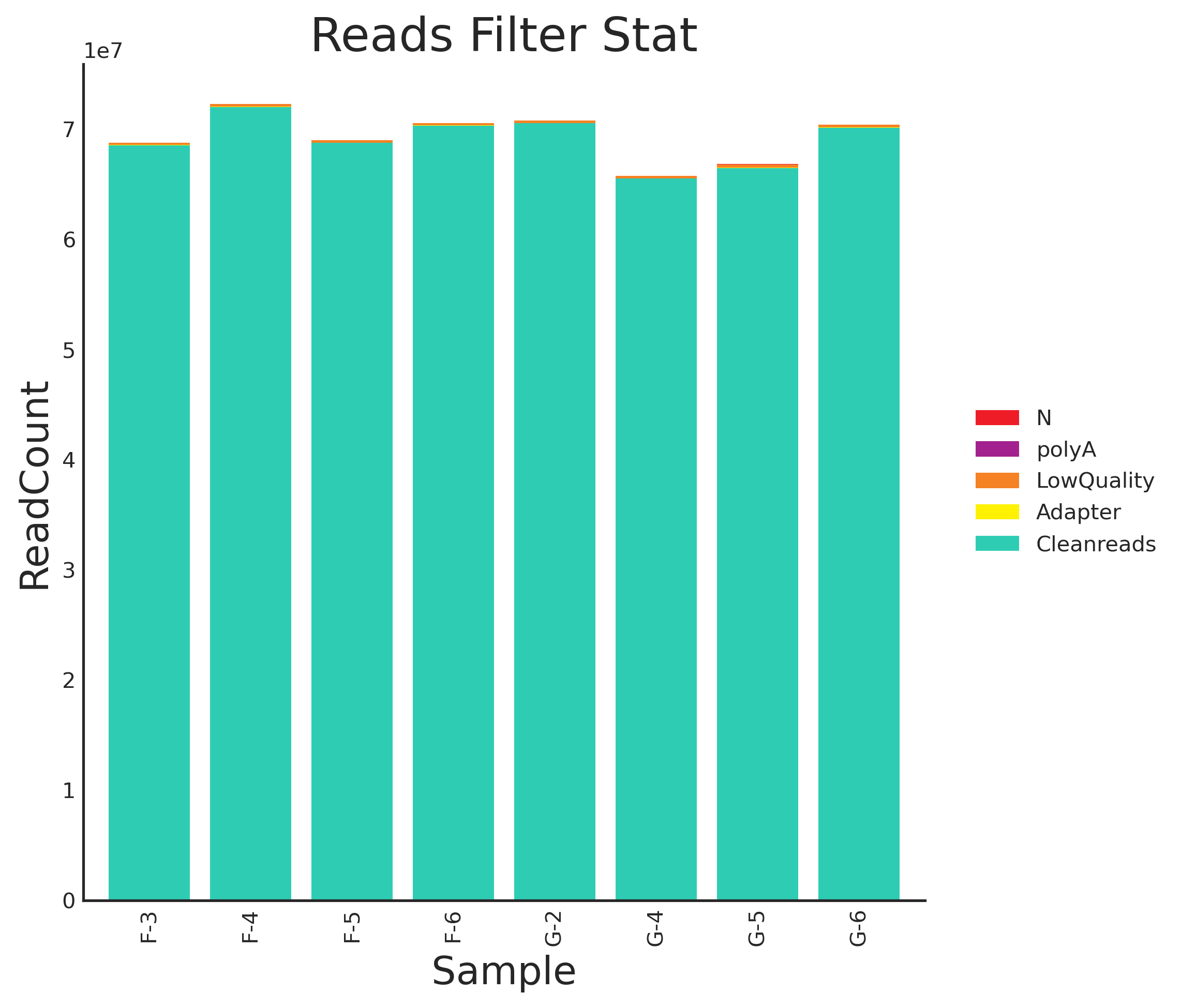
Tab 3-1-1 数据过滤统计表
| Sample | Rawreads | Cleanreads(%) | Adapter(%) | LowQuality(%) | polyA(%) | N(%) |
|---|
| F-3 | 68663090 | 68526930 (99.80%) | 30706 (0.04%) | 207064 (0.15%) | 0 (0.0%) | 3844 (0.0%) |
| F-4 | 72155062 | 72003126 (99.79%) | 31220 (0.04%) | 231280 (0.16%) | 0 (0.0%) | 10152 (0.01%) |
| F-5 | 68876222 | 68756650 (99.83%) | 22020 (0.03%) | 191168 (0.14%) | 0 (0.0%) | 3936 (0.0%) |
| F-6 | 70430554 | 70294040 (99.81%) | 35404 (0.05%) | 192124 (0.14%) | 0 (0.0%) | 10096 (0.01%) |
| G-2 | 70665654 | 70529454 (99.81%) | 29390 (0.04%) | 200644 (0.14%) | 0 (0.0%) | 12976 (0.01%) |
| G-4 | 65631282 | 65497078 (99.80%) | 25712 (0.04%) | 213268 (0.16%) | 0 (0.0%) | 3716 (0.0%) |
| G-5 | 66654320 | 66466480 (99.72%) | 32438 (0.05%) | 307528 (0.23%) | 0 (0.0%) | 3276 (0.0%) |
| G-6 | 70276890 | 70103678 (99.75%) | 39056 (0.06%) | 257332 (0.18%) | 0 (0.0%) | 10980 (0.01%) |
![]() |
|
![]() |
Fig 3-1-1 数据预处理分布图(百分比) |
|
Fig 3-1-2 数据预处理分布图(数值) |
|
|
|
3.2 碱基质量分析
数据经过过滤后,我们将分析碱基的组成及质量分布,以直观展示数据质量情况。碱基组成越平衡,质量越高,后续分析则越准确。
Tab 3-2-1 过滤前后碱基信息统计表
| Sample | RawData(bp) | BF_Q20(%) | BF_Q30(%) | BF_N(%) | BF_GC(%) | CleanData(bp) | AF_Q20(%) | AF_Q30(%) | AF_N(%) | AF_GC(%) |
|---|
| F-3 | 10299463500 | 9993944910 (97.03%) | 9465326967 (91.9%) | 193188 (0.0%) | 5216314828 (50.64%) | 10261977261 | 9964697107 (97.1%) | 9439740968 (91.99%) | 164746 (0.0%) | 5195899500 (50.64%) |
| F-4 | 10823259300 | 10468860149 (96.73%) | 9878770979 (91.27%) | 323304 (0.0%) | 5410541082 (49.99%) | 10791132428 | 10445617444 (96.8%) | 9859139775 (91.36%) | 172924 (0.0%) | 5393133246 (49.98%) |
| F-5 | 10331433300 | 9995149257 (96.75%) | 9429336703 (91.27%) | 195341 (0.0%) | 5320552875 (51.5%) | 10301616881 | 9972739741 (96.81%) | 9410164154 (91.35%) | 166477 (0.0%) | 5304212894 (51.49%) |
| F-6 | 10564583100 | 10230573448 (96.84%) | 9666715357 (91.5%) | 323923 (0.0%) | 5151257722 (48.76%) | 10532195019 | 10207147983 (96.91%) | 9646949221 (91.59%) | 172239 (0.0%) | 5134097258 (48.74%) |
| G-2 | 10599848100 | 10302104514 (97.19%) | 9776550923 (92.23%) | 380456 (0.0%) | 5446545493 (51.38%) | 10566414210 | 10276029120 (97.25%) | 9753679888 (92.31%) | 172419 (0.0%) | 5428627622 (51.38%) |
| G-4 | 9844692300 | 9505992364 (96.56%) | 8953934569 (90.95%) | 186818 (0.0%) | 5066197778 (51.46%) | 9812233057 | 9481993083 (96.63%) | 8933606112 (91.05%) | 159546 (0.0%) | 5048577787 (51.45%) |
| G-5 | 9998148000 | 9654188365 (96.56%) | 9095462182 (90.97%) | 180539 (0.0%) | 5127872541 (51.29%) | 9964204920 | 9630488454 (96.65%) | 9075754142 (91.08%) | 155924 (0.0%) | 5109396306 (51.28%) |
| G-6 | 10541533500 | 10241841278 (97.16%) | 9718340579 (92.19%) | 338064 (0.0%) | 5416351405 (51.38%) | 10492366443 | 10202518463 (97.24%) | 9683481247 (92.29%) | 170359 (0.0%) | 5389651322 (51.37%) |
- F-3
- F-4
- F-5
- F-6
- G-2
- G-4
- G-5
- G-6
Fig 3-2-1 各样品过滤前后碱基组成分布图
3.3 宿主序列过滤
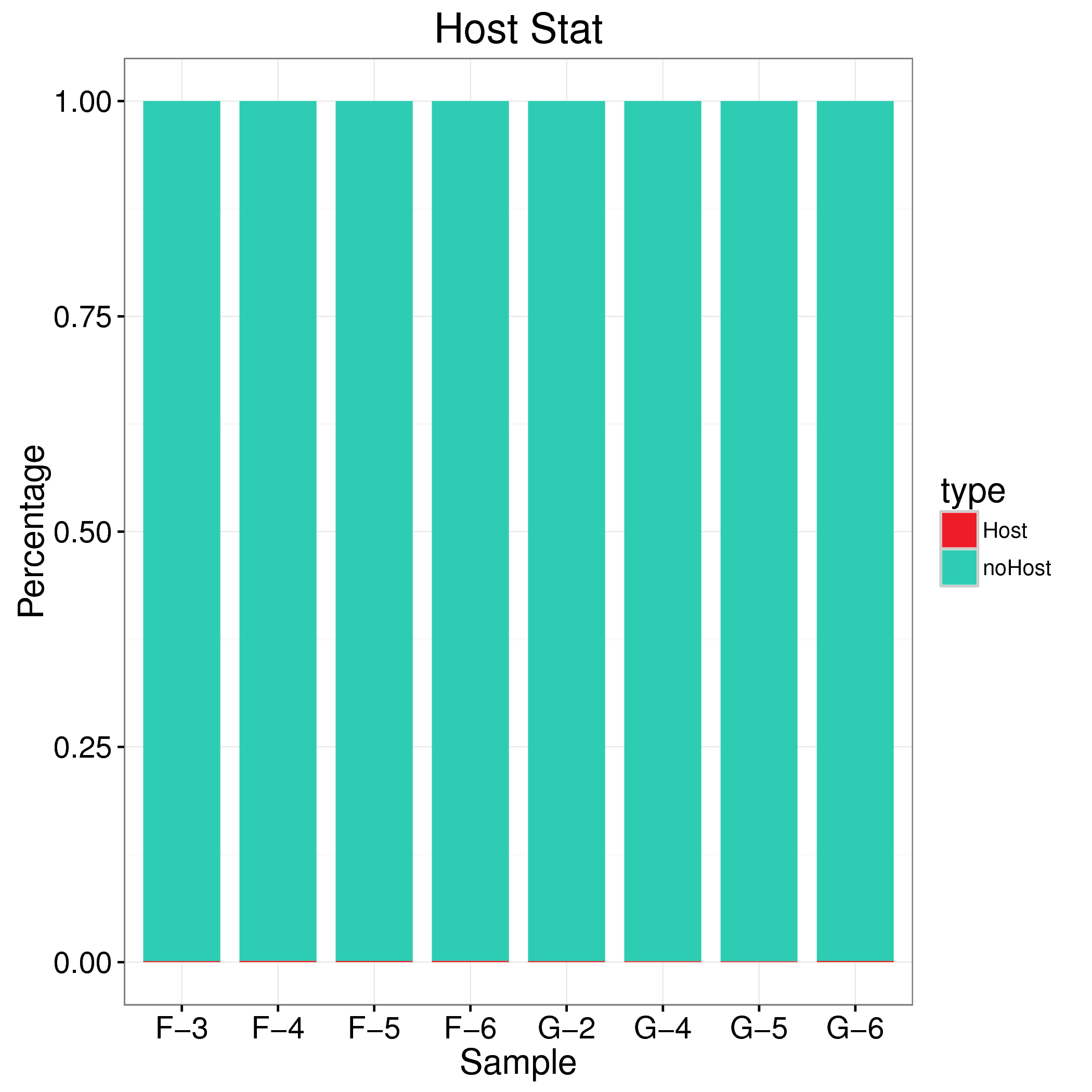
如果样本采集来源于肠道等微生物,则不可避免的会存在宿主序列污染。如果宿主参考基因组已经发布,我们会将过滤后数据用 Bowtie2 比对到宿主参考基因组,过滤来源于宿主的reads,得到effective reads进行后续分析。
备注:此分析只在样本存在宿主参考的情况下才会开展。
Tab 3-3-1 宿主序列过滤统计表
| Samples | F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 |
|---|
| Sum Clean Reads | 68526930 | 72003126 | 68756650 | 70294040 | 70529454 | 65497078 | 66466480 | 70103678 |
| Sum Host mapped Reads | 89355 | 110297 | 97751 | 111396 | 92941 | 78777 | 74682 | 104773 |
![]() |
| Fig 3-3-1 宿主序列过滤统计 |
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
4 序列组装
利用 MEGAHIT 软件对effective reads进行组装
结果在文件夹:02.Assemble下
Tab 4-0-1 各样本组装统计结果
| Sample | Contigs Num | Total length | Average length | Max length | N50 | N90 |
|---|
| F | 1361185 | 2184597799 | 1604.92 | 619133 | 2326 | 645 |
| G | 1576316 | 2206043563 | 1399.49 | 331676 | 1748 | 616 |
- 样品 F contig 长度分布图
- 样品 G contig 长度分布图
Fig 4-0-1 各样本contig长度分布图
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
5 基因预测
5.1 基因预测
利用 MetaGeneMark 对>500bp的contigs进行基因预测,然后采用 CD-HIT 软件(95% identity、90% coverage)对所预测基因进行聚类,选取最长的基因作为每类代表序列,构建初始非冗余基因集合。
结果在文件夹:03.Genes下
Tab 5-1-1 非冗余基因集样本基因数目统计表
| Sample | GeneNumber | TotalLength | AverageLength | GC% |
|---|
| F-3 | 461162 | 515203104 | 1117 | 53.79% |
| F-4 | 537898 | 580964919 | 1080 | 53.63% |
| F-5 | 400314 | 460464543 | 1150 | 53.70% |
| F-6 | 395609 | 452996859 | 1145 | 53.30% |
| G-2 | 388715 | 427116321 | 1098 | 54.20% |
| G-4 | 293558 | 353490087 | 1204 | 54.12% |
| G-5 | 415056 | 460258407 | 1108 | 54.58% |
| G-6 | 567078 | 576597696 | 1016 | 54.10% |
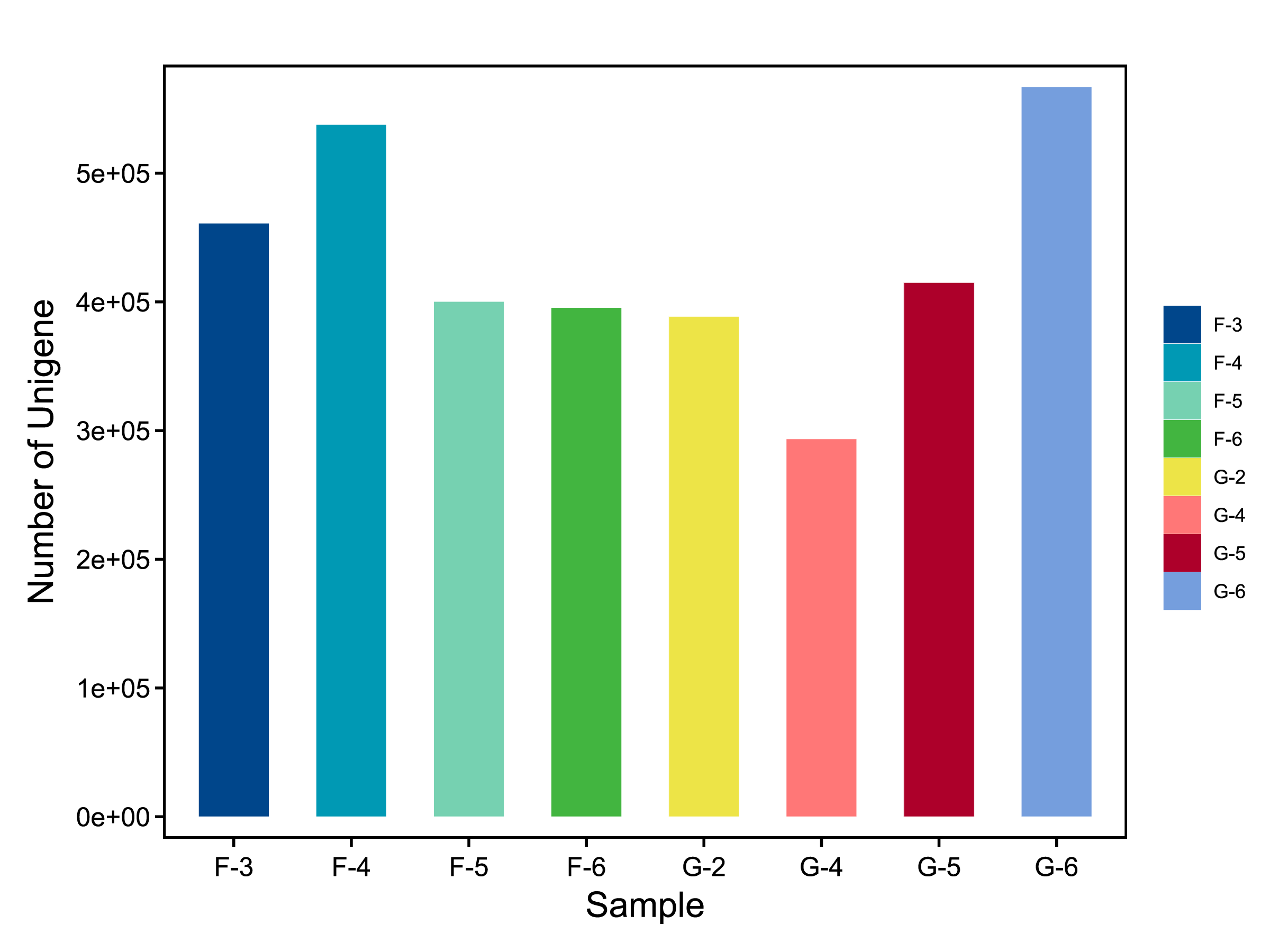
使用柱形图展示每个样本的基因数目分布,便于比较样本间基因数量差异。
![]() |
Fig 5-1-1 各样本基因数目分布 |
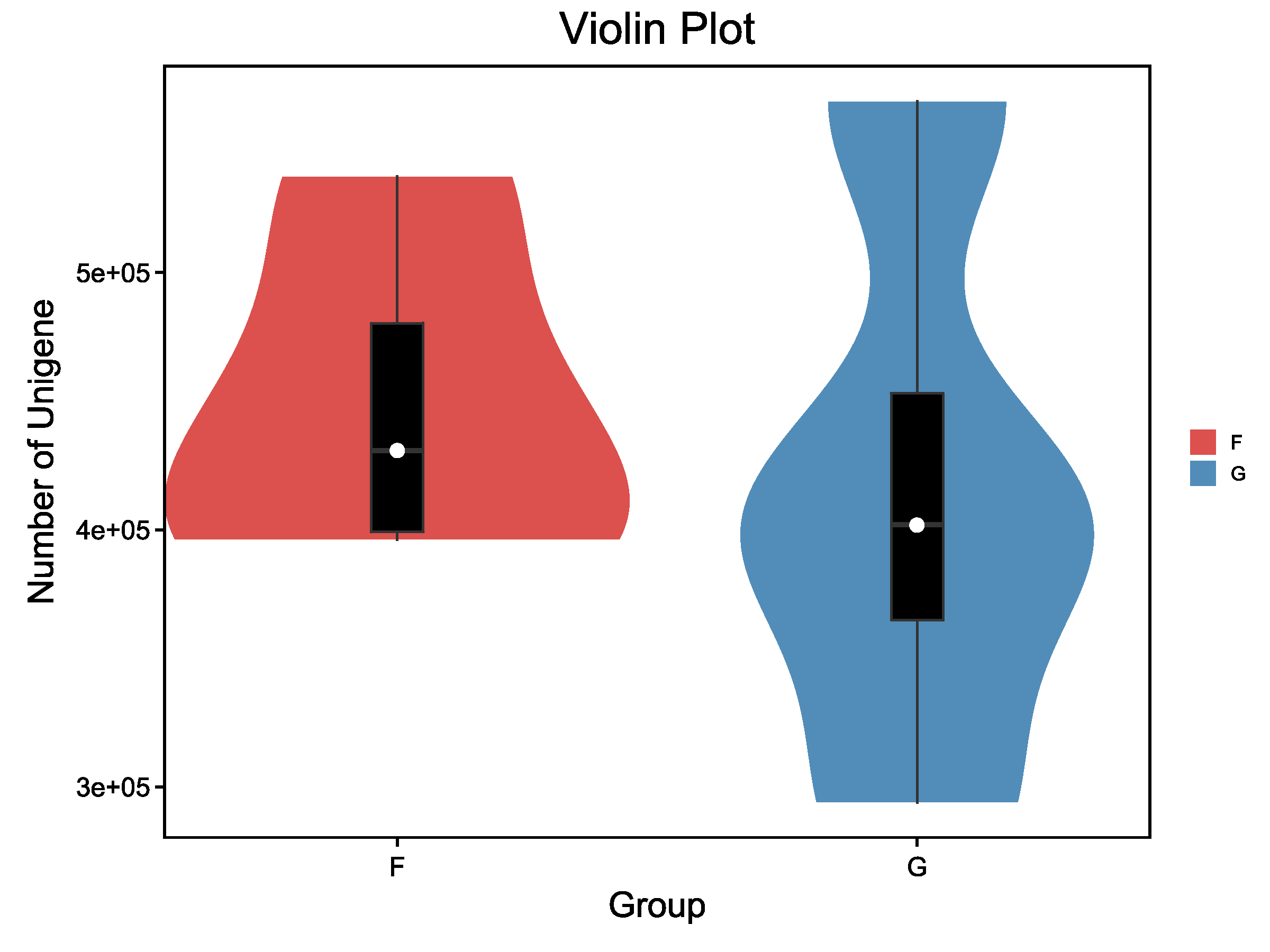
使用小提琴图可直观反映组内样本基因数目分布和组间基因数目的差异。
![]() |
Fig 5-1-2 分组基因数分布小提琴图 |
5.2 基因丰度统计
利用 bowtie2 将 clean reads重新比对到初始非冗余基因集上,并基于比对结果,使用pathoscope软件重新将reads分配给最佳基因。(备注:Bowtie比对时可能出现一条reads比对上多个基因,而pathoscope可以基于算法将这些多重比对的reads分配给最佳基因,优化基因定量结果,也是文献认可推荐的方法)。过滤掉在各个样品中reads支持数目≤2的基因,获得最终用于后续分析的基因集合。
从分配给基因的reads 数目、基因长度、测序深度出发,计算得到各基因在各样品中的相对丰度信息。前10行示例如下:
Tab 5-2-1 各样本基因丰度表
| GeneID | F-3_count | F-4_count | F-5_count | F-6_count | G-2_count | G-4_count | G-5_count | G-6_count | F-3_RelativeAbundance | F-4_RelativeAbundance | F-5_RelativeAbundance | F-6_RelativeAbundance | G-2_RelativeAbundance | G-4_RelativeAbundance | G-5_RelativeAbundance | G-6_RelativeAbundance |
|---|
| Unigene1 | 13.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9.038851272516904e-07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene4 | 0 | 0 | 0 | 3.0 | 0 | 0 | 0 | 16.0 | 0 | 0 | 0 | 2.527775465885085e-07 | 0 | 0 | 0 | 1.5335500503915718e-06 |
| Unigene13 | 0 | 0 | 0 | 283.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.4873831280945607e-05 | 0 | 0 | 0 | 0 |
| Unigene14 | 18.0 | 0 | 7.0 | 0 | 0 | 0 | 0 | 0 | 8.871162176510935e-07 | 0 | 3.363301974977882e-07 | 0 | 0 | 0 | 0 | 0 |
| Unigene16 | 0 | 0 | 3.0 | 4.0 | 0 | 30.0 | 0 | 0 | 0 | 0 | 1.8493628110390513e-07 | 2.4037713864265717e-07 | 0 | 2.967263527620739e-06 | 0 | 0 |
| Unigene17 | 11.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.155886563458923e-07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene18 | 0 | 15.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.5778178350403477e-06 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene19 | 0 | 0 | 0 | 28.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.1181789663085946e-06 | 0 | 0 | 0 | 0 |
| Unigene20 | 0 | 0 | 0 | 16.0 | 0 | 0 | 0 | 4.0 | 0 | 0 | 0 | 1.6545439413066012e-06 | 0 | 0 | 0 | 4.705210381883232e-07 |
| Unigene22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
结果在文件夹:03.Genes
- 基因CDS序列:Unigenes.final.fna
- 基因编码蛋白序列:Unigenes.final.faa
- 基因在各个样本中的丰度:Unigenes.expression.final.xls
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
6 基因功能注释
获得非冗余基因集后,我们基于各种数据库比对注释结果分析预测样本中微生物群落的功能特征。将Unigenes通过DIAMOND软件(阈值evalue<=1e-5)比对到KEGG、eggNOG、CAZy、CARD、VFDB、PHI等多个数据库,同时集合基因丰度表格计算不同数据库比对结果的丰度信息,以进行系统丰富的组间功能差异分析和比较。
结果在文件夹:04.Annotation下
Tab 6-0-1 数据库注释统计表
| Database | GeneCount | GenePercent(%) |
|---|
| KEGG | 1633079 | 71.92% |
| eggNOG | 1676538 | 73.83% |
| CAZy | 271550 | 11.96% |
| VFDB | 203451 | 8.96% |
| PHI | 231736 | 10.20% |
| CARD | 63610 | 2.80% |
6.1 KEGG功能注释
KEGG,全称Kyoto Encyclopedia of Genes and Genomes,是一个关于基因功能注释方面的综合性数据库,包括基因的功能、分类、代谢通路(KEGG Pathway数据库, 是KEGG最核心的功能注释数据库)等诸多方面的信息。
KEGG Pathway数据库将生物代谢通路划分为7大类(A级分类,level 1),分别为:新陈代谢(Metabolism)、遗传信息处理(Genetic Information Processing)、 环境信息处理(Environmental Information Processing)、细胞过程(Cellular Processes)、生物体系统(Organismal Systems)、 人类疾病(Human Diseases)、药物相关代谢(Drug developmennt)。每大类又被逐步细分更具体的B、C、D3个层级。目前B级分类(level 2)共有 59个子分类。C级分类(level 3)即为代谢通路图(pathway,map);D级分类为每个代谢通路图中具体的酶、同源基因、化合物等信息。
我们基于KEGG的层级分类注释结果,可以获得样本/分组间不同深度的功能特征,还可以基于pathway注释信息系统了解群落内基因间的潜在关联、代谢网络等。
Pathway注释信息展示如下:
Tab 6-1-1 Pathway 注释信息
| KEGG_A_class | KEGG_B_class | Pathway | Count (453974) | Pathway ID | ... |
|---|
| Metabolism | Nucleotide metabolism | Purine metabolism | 37930 | ko00230 | ... |
| Environmental Information Processing | Membrane transport | ABC transporters | 33855 | ko02010 | ... |
| Metabolism | Nucleotide metabolism | Pyrimidine metabolism | 33616 | ko00240 | ... |
| Environmental Information Processing | Signal transduction | Two-component system | 27270 | ko02020 | ... |
| Cellular Processes | Cellular community - prokaryotes | Quorum sensing | 24892 | ko02024 | ... |
| Metabolism | Carbohydrate metabolism | Amino sugar and nucleotide sugar metabolism | 24328 | ko00520 | ... |
| Metabolism | Carbohydrate metabolism | Starch and sucrose metabolism | 23047 | ko00500 | ... |
| Genetic Information Processing | Replication and repair | Homologous recombination | 20545 | ko03440 | ... |
| Genetic Information Processing | Translation | Aminoacyl-tRNA biosynthesis | 20316 | ko00970 | ... |
| Genetic Information Processing | Replication and repair | Mismatch repair | 17826 | ko03430 | ... |
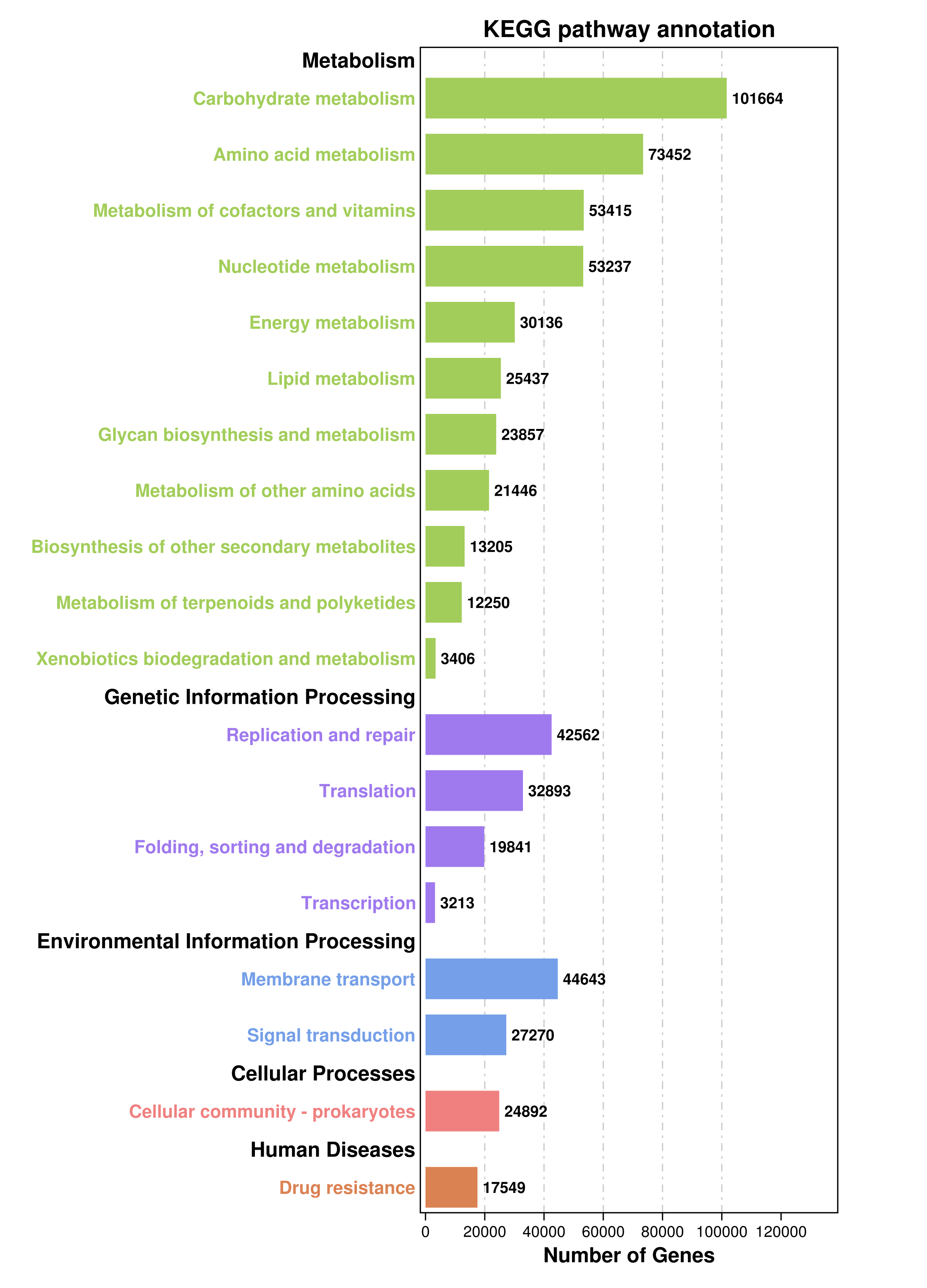
我们统计所有样本中注释到pathway数据库的基因总数,并绘制条形图,直观展示不同分类层级群落的基因数量分布情况
![]() |
Fig 6-1-1 所有样本中pathway注释到的基因数目 |
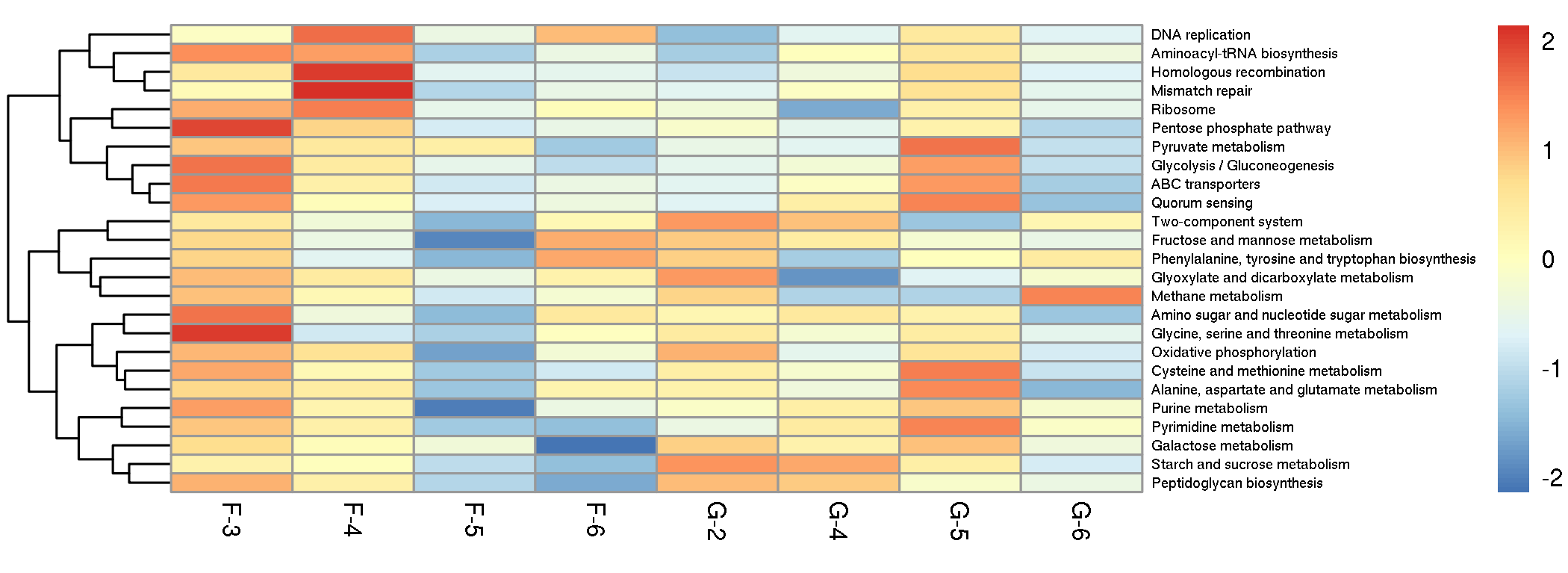
我们用热图展示各样本中KEGG不同层级的功能丰度特征,直观呈现每个样本的功能丰度信息,以初步呈现样本/分组间功能分布规律。并挑选pathway(所有样本中丰度之和排名前25的pathway)丰度特征展示如下。
Tab 6-1-2 Pathway热图数据
| F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 |
|---|
| Purine metabolism | 0.0181247013946175 | 0.0173439634866131 | 0.0155784236035765 | 0.0168113749268161 | 0.0170730271803261 | 0.0174283269256638 | 0.0178797398374333 | 0.0170129829617836 |
| ABC transporters | 0.0216879186096565 | 0.017719030333573 | 0.0139750642108909 | 0.0152078289053535 | 0.0147521945807893 | 0.0164334131359709 | 0.0209603513471051 | 0.0128013425517915 |
| Pyrimidine metabolism | 0.0156549971688603 | 0.0152405604441294 | 0.0142156337550766 | 0.0141317698204458 | 0.0147426504598997 | 0.0153385920687924 | 0.0160229713526845 | 0.0149757073078571 |
| Two-component system | 0.0141834815288719 | 0.0135596410702269 | 0.0126581293817935 | 0.0139494568247399 | 0.0148469393661482 | 0.0145652379587045 | 0.0128025922261919 | 0.0139700568239606 |
| Quorum sensing | 0.0148030563841526 | 0.0123941626535127 | 0.0108704365317802 | 0.0114904366219596 | 0.0109787689533888 | 0.012975702607063 | 0.0151468057773745 | 0.00964859975212261 |
| Starch and sucrose metabolism | 0.0113180828067974 | 0.0109529337404188 | 0.00949091207660668 | 0.00899768096483018 | 0.0128198532111878 | 0.012592884517725 | 0.0114581918458801 | 0.00982237296609164 |
| Amino sugar and nucleotide sugar metabolism | 0.0115809978830278 | 0.0104193172822981 | 0.00978633999817393 | 0.0109361213655275 | 0.0107510942910294 | 0.0109185187284085 | 0.0108092671117915 | 0.0098602318609377 |
| Homologous recombination | 0.0100411245300785 | 0.0114945471402651 | 0.00898117641981112 | 0.00901871879937547 | 0.00869768808876349 | 0.00920689021594226 | 0.0102429320884549 | 0.00887958423605778 |
| Aminoacyl-tRNA biosynthesis | 0.00950765076955259 | 0.00943522490667251 | 0.00809035132967498 | 0.00849156694553213 | 0.00805816756143655 | 0.00876407220305832 | 0.00904611762657461 | 0.00854373607515917 |
| Ribosome | 0.0100839820365126 | 0.0106362782561463 | 0.0074462889747173 | 0.00834741570468766 | 0.00775814302749779 | 0.0057439416569913 | 0.00877884563849209 | 0.00743489280176606 |
![]() |
Fig 6-1-2 Top 25 pathway Heatmap |
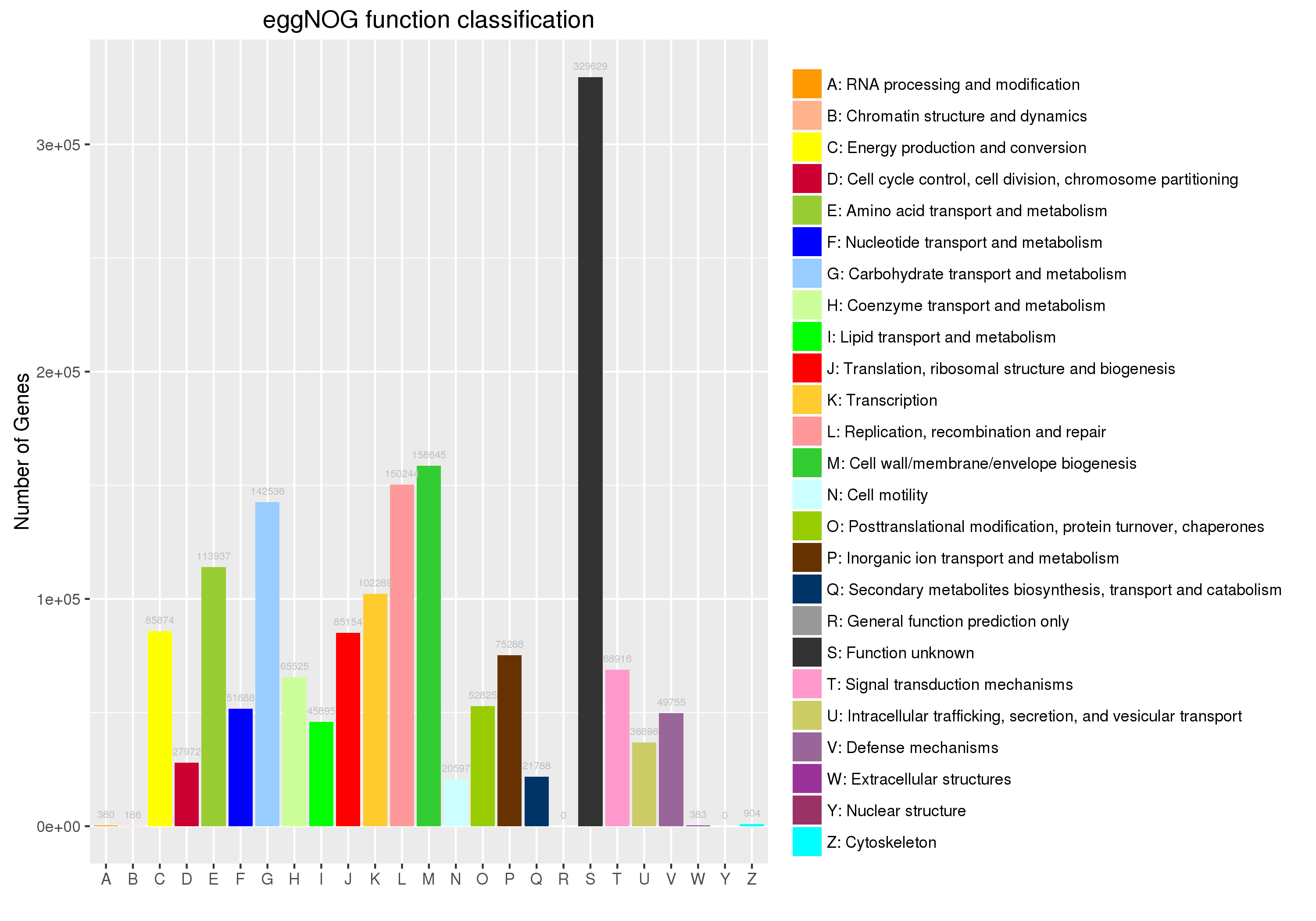
6.2 eggNOG功能注释
eggNOG(evolutionary genealogy of genes: Non-supervised Orthologous Groups)数据库是利用 Smith-Waterman 比对算法构建的基因直系同源簇 (Orthologous Groups,Ogs),当前最新版本5.0(2019.01),涵盖了5090个物种(4445个代表性细菌、168个古菌、477个真核生物)、2502个病毒,首先基于所有基因组蛋白信息,聚类获得约4.4M的OGs(C层级),然后基于KEGG等各类数据库进行注释,最终构建成25个功能分类(A层级)。
使用eggNOG数据库基于直系同源(orthology)进行基因、蛋白序列的功能预测被认为比传统的同源搜索更准确,常应用于新基因组、宏基因组等基因集。我们使用Diamond进行eggNOG注释,并以柱形图展示25个功能分类的基因数目统计情况,便于对比群落中各功能分类的基因数量分布。
Tab 6-2-1 eggNOG注释详细信息
| GeneID | eggNOG_OG | Best_Description | Best_COG_Cat | F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 |
|---|
| Unigene1 | COG0345;1TP1E;27JFY;247SR | Catalyzes the reduction of 1-pyrroline-5-carboxylate (PCA) to L-proline | E | 9.038851272516904e-07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene13 | COG0577;2FPEN;4NDUK;22X01 | ABC transporter permease | V | 0 | 0 | 0 | 1.4873831280945607e-05 | 0 | 0 | 0 | 0 |
| Unigene14 | 2J5C6;COG0527;COG0460 | Amino acid kinase family | E | 8.871162176510935e-07 | 0 | 3.363301974977882e-07 | 0 | 0 | 0 | 0 | 0 |
| Unigene16 | COG2217 | Heavy metal translocating P-type atpase | P | 0 | 0 | 1.8493628110390513e-07 | 2.4037713864265717e-07 | 0 | 2.967263527620739e-06 | 0 | 0 |
| Unigene17 | 4E7E2;2GQPQ;COG0681 | Peptidase S24-like | U | 8.155886563458923e-07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene19 | 1TNYD;247QZ;COG1126;4BXFR | ATPases associated with a variety of cellular activities | E | 0 | 0 | 0 | 3.1181789663085946e-06 | 0 | 0 | 0 | 0 |
| Unigene20 | COG3152 | Membrane | L | 0 | 0 | 0 | 1.6545439413066012e-06 | 0 | 0 | 0 | 4.705210381883232e-07 |
| Unigene22 | 1ZBDP;COG0557;1TQ1G;4HBBH | 3'-5' exoribonuclease that releases 5'-nucleoside monophosphates and is involved in maturation of structured RNAs | K | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene37 | 33MU2;24W74;1VQ3S;2DTVE | - | - | 0 | 8.560410042685029e-07 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene44 | 247XY;27J0H;COG0090;1TP9X | One of the primary rRNA binding proteins. Required for association of the 30S and 50S subunits to form the 70S ribosome, for tRNA binding and peptide bond formation. It has been suggested to have peptidyltransferase activity | J | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
![]() |
Fig 6-2-1 eggNOG注释统计图 |
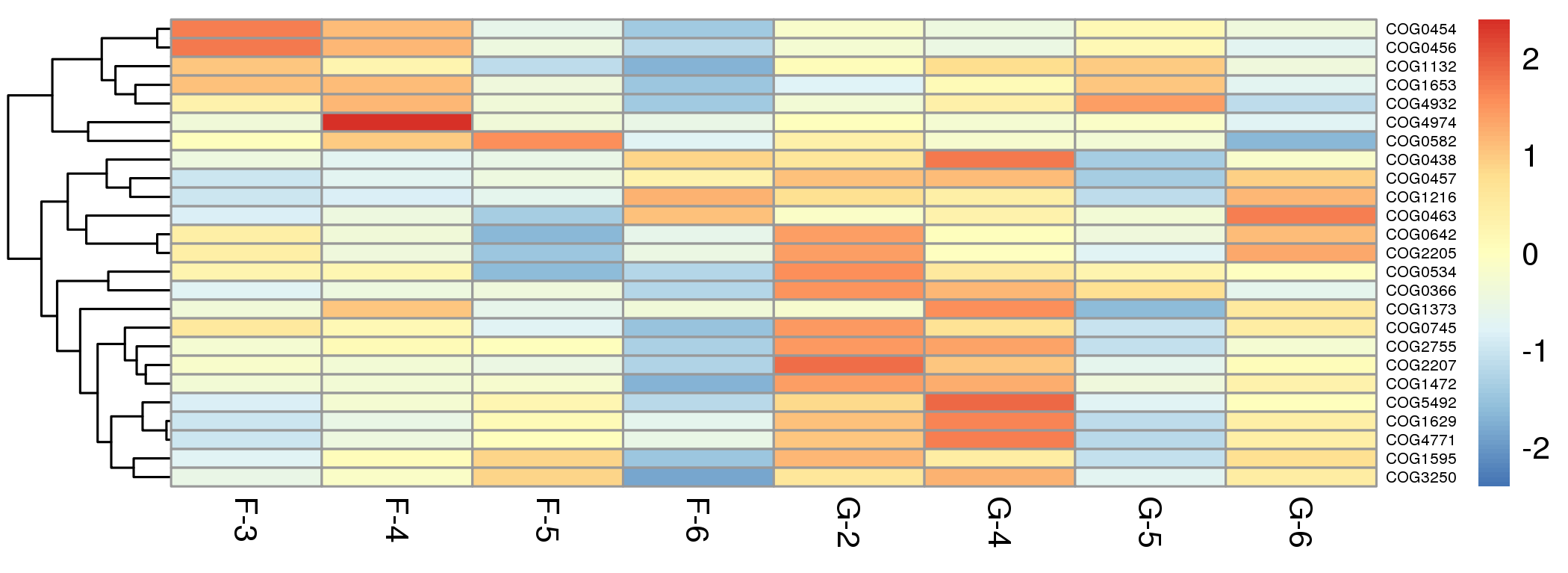
热图展示总丰度排名前25的Ogs在所有样本中的丰度分布特征,便于初步查看各样本/分组功能分布规律。
![]() |
Fig 6-2-2 Top 25 eggNOG Family Heatmap |
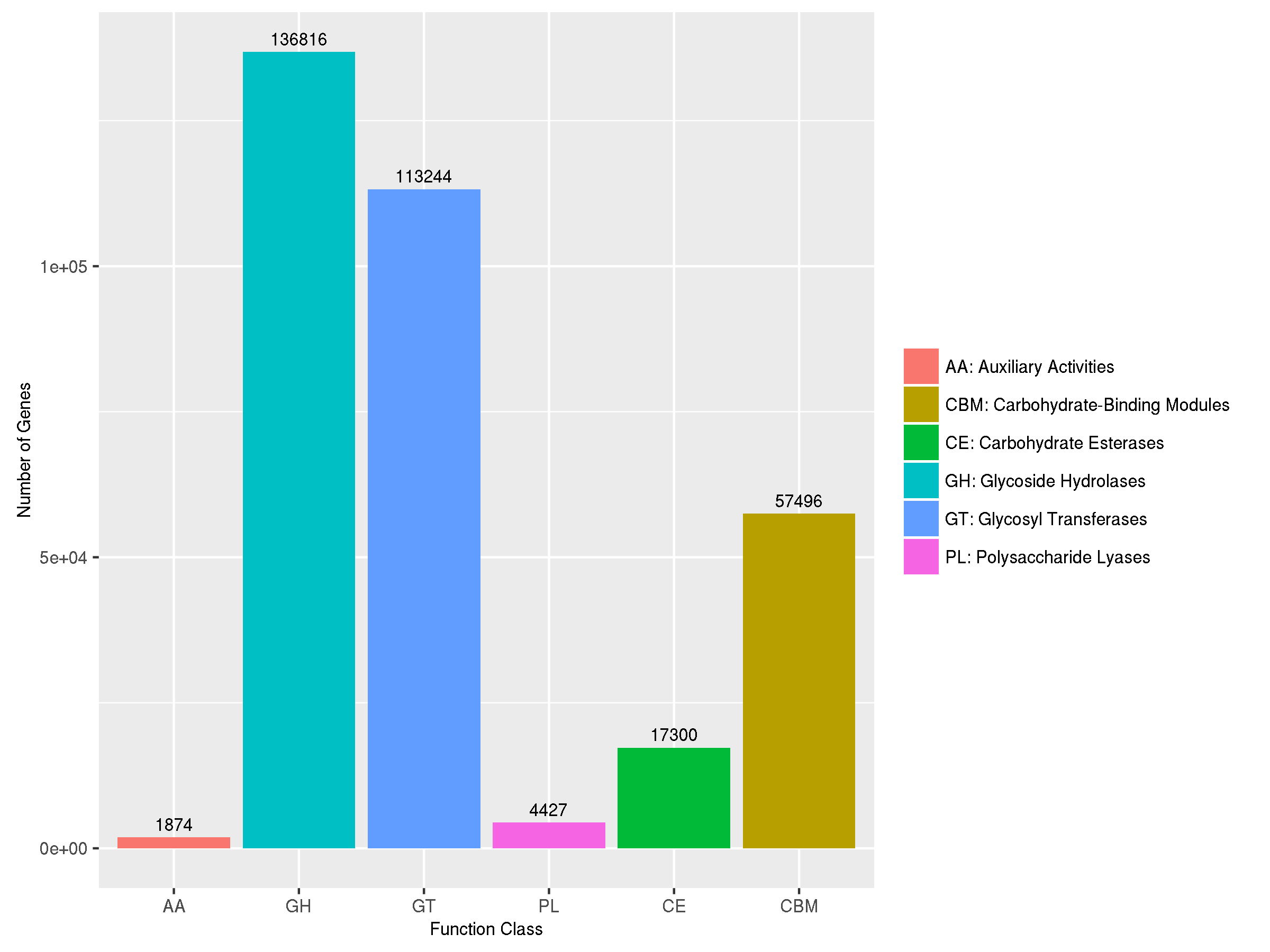
6.3 CAZy功能注释
CAZy 全称Carbohydrate-Active enZYmes Database,是碳水化合物酶相关的专业数据库,包括催化碳水化合物生物合成、降解以及修饰的相关酶系家族,对生物质转化等工业微生物领域、生境内碳循环相关功能研究有重要参考价值。
其包含6个主要分类(level A):糖苷水解酶(Glycoside Hydrolases, GHs)、糖基转移酶(Glycosyl Transferases, GTs)、多糖裂解酶(Polysaccharide Lyases, PLs)和糖类酯解酶(Carbohydrate Esterases, CEs)、辅助氧化还原酶(Auxiliary Activities , AAs)。此外,还包含与碳水化合物相关的modules(Carbohydrate-Binding Modules,CBMs)。其中每一个大类有可以分类很多小的家族(level B),比如CE1,CE2等等,注释结果中的CE0表示没有小家族分类的结果。
使用 Diamond 将基因序列与数据库比对注释。
Tab 6-3-1 CAZy注释详细信息
| LevelA | LevelA_full_name | LevelB | Activities | Count | Genes |
|---|
| AA | Auxiliary Activities | AA0 | - | 171 | Unigene2417013;Unigene4778820;Unigene246502;Unigene833634;Unigene142581;Unigene916912;Unigene3839417;Unigene317618;Unigene4380675;Unigene3067376;Unigene328007;Unigene368754;Unigene691405;Unigene3059373;Unigene4897004;Unigene4894757;Unigene606610;Unigene2468769;Unigene368637;Unigene115203;Unigene1828892;Unigene4209982;Unigene2247671;Unigene1564168;Unigene2820379;Unigene4403997;Unigene2087114;Unigene2034919;Unigene2490462;Unigene725027;Unigene1596940;Unigene3984534;Unigene3301920;Unigene1326868;Unigene3502815;Unigene3567017;Unigene2614586;Unigene1972655;Unigene2070955;Unigene2371071;Unigene4796683;Unigene2806240;Unigene3000077;Unigene134863;Unigene402242;Unigene1316062;Unigene2443483;Unigene843210;Unigene3958112;Unigene143576;Unigene745049;Unigene2650200;Unigene217947;Unigene3306214;Unigene3361296;Unigene2857693;Unigene3611334;Unigene3642107;Unigene1094225;Unigene2700547;Unigene306101;Unigene3019758;Unigene1393796;Unigene1458813;Unigene620060;Unigene1711777;Unigene3638039;Unigene1308691;Unigene1736090;Unigene3243624;Unigene1801610;Unigene4922574;Unigene1421439;Unigene568012;Unigene1168374;Unigene3589332;Unigene2272739;Unigene3389935;Unigene1467990;Unigene4646187;Unigene2206278;Unigene110666;Unigene4753258;Unigene1759591;Unigene682281;Unigene852983;Unigene4944682;Unigene4762264;Unigene1419572;Unigene3638242;Unigene478735;Unigene55173;Unigene4816488;Unigene4090779;Unigene1108793;Unigene210682;Unigene945538;Unigene2842192;Unigene502881;Unigene844310;Unigene2317398;Unigene2761303;Unigene1973477;Unigene67928;Unigene2047526;Unigene4333266;Unigene3034547;Unigene3844637;Unigene2613560;Unigene155801;Unigene1134610;Unigene211071;Unigene4402777;Unigene1715536;Unigene3957792;Unigene4904180;Unigene3424598;Unigene3854360;Unigene4069762;Unigene1235721;Unigene1084220;Unigene4146426;Unigene1579755;Unigene237597;Unigene1711780;Unigene805712;Unigene2975806;Unigene1273474;Unigene4398328;Unigene655064;Unigene3832025;Unigene4803528;Unigene160764;Unigene4288945;Unigene1719210;Unigene49538;Unigene1777645;Unigene972542;Unigene3842619;Unigene101965;Unigene354799;Unigene1737327;Unigene4646331;Unigene546618;Unigene1923986;Unigene3952161;Unigene4730928;Unigene4625935;Unigene3268823;Unigene1552035;Unigene4843696;Unigene4832689;Unigene2073225;Unigene2138771;Unigene3886949;Unigene1899510;Unigene1950996;Unigene1222079;Unigene633048;Unigene4506458;Unigene1905972;Unigene1003257;Unigene4099943;Unigene1480089;Unigene3592139;Unigene1629245;Unigene2529922;Unigene3511261;Unigene3256265;Unigene2244553;Unigene4009537 |
| AA | Auxiliary Activities | AA1 | Laccase / p-diphenol:oxygen oxidoreductase / ferroxidase (EC 1.10.3.2); ; ferroxidase (EC 1.10.3.-); Laccase-like multicopper oxidase (EC 1.10.3.-) | 1340 | Unigene4494904;Unigene2650398;Unigene109960;Unigene1595068;Unigene3387598;Unigene217586;Unigene2379411;Unigene1460983;Unigene2399477;Unigene1911754;Unigene2635994;Unigene387835;Unigene3477056;Unigene2844139;Unigene2832383;Unigene4304991;Unigene4501005;Unigene253985;Unigene1485511;Unigene3336199;Unigene4748107;Unigene1463220;Unigene3480967;Unigene2355287;Unigene3875581;Unigene2434751;Unigene4245199;Unigene4694710;Unigene3551672;Unigene537855;Unigene290631;Unigene4130369;Unigene4299887;Unigene1666324;Unigene362023;Unigene2878761;Unigene4021706;Unigene4329;Unigene2661212;Unigene4402177;Unigene3756402;Unigene3012241;Unigene2251939;Unigene4735655;Unigene665704;Unigene630508;Unigene3744199;Unigene1420447;Unigene1240468;Unigene3693717;Unigene1513208;Unigene3516185;Unigene946701;Unigene2717828;Unigene71417;Unigene3566059;Unigene1090300;Unigene1096005;Unigene3128241;Unigene424910;Unigene2919977;Unigene3095034;Unigene2088920;Unigene2941628;Unigene918055;Unigene28869;Unigene103815;Unigene2114584;Unigene1859574;Unigene820788;Unigene844105;Unigene3627710;Unigene3024214;Unigene4757114;Unigene1170715;Unigene3546555;Unigene206537;Unigene3607501;Unigene4440215;Unigene4339078;Unigene4753594;Unigene3328575;Unigene64371;Unigene1040502;Unigene3024784;Unigene2108827;Unigene4508782;Unigene3376344;Unigene17262;Unigene43688;Unigene1407532;Unigene3075855;Unigene605246;Unigene4167177;Unigene3177563;Unigene1185303;Unigene4356617;Unigene1640118;Unigene4583837;Unigene3739161;Unigene693562;Unigene1865518;Unigene2000996;Unigene4281161;Unigene1022087;Unigene345851;Unigene1319312;Unigene43964;Unigene1077501;Unigene2733004;Unigene3520293;Unigene1833160;Unigene818192;Unigene3602451;Unigene3204235;Unigene527734;Unigene2908343;Unigene4084609;Unigene1299267;Unigene420865;Unigene3771942;Unigene1719151;Unigene2997691;Unigene2644273;Unigene3912460;Unigene2077689;Unigene4867915;Unigene1715527;Unigene2891130;Unigene165010;Unigene2244776;Unigene916966;Unigene365348;Unigene2169862;Unigene3450392;Unigene2374365;Unigene1688345;Unigene4187345;Unigene1074436;Unigene4325804;Unigene4386949;Unigene224901;Unigene2681722;Unigene2101431;Unigene1969778;Unigene1422248;Unigene1141211;Unigene2567;Unigene917747;Unigene2719602;Unigene3484943;Unigene3542843;Unigene2393396;Unigene4696534;Unigene3873544;Unigene1644328;Unigene1000235;Unigene4587059;Unigene144260;Unigene462427;Unigene4905580;Unigene2575532;Unigene4735510;Unigene951591;Unigene3158591;Unigene1417771;Unigene414140;Unigene718315;Unigene1681641;Unigene3857042;Unigene1438598;Unigene4203831;Unigene4078539;Unigene425913;Unigene1465848;Unigene2268451;Unigene2397087;Unigene611685;Unigene1801693;Unigene219604;Unigene4093384;Unigene98047;Unigene1716438;Unigene4581348;Unigene3865184;Unigene2357716;Unigene3938577;Unigene1882215;Unigene2126165;Unigene4298537;Unigene856284;Unigene36001;Unigene923552;Unigene3076166;Unigene2024005;Unigene1495644;Unigene1947136;Unigene1287387;Unigene3019898;Unigene1755033;Unigene614395;Unigene4713268;Unigene2820117;Unigene3636644;Unigene4524447;Unigene4361858;Unigene4696217;Unigene451891;Unigene4298660;Unigene4860877;Unigene651140;Unigene1255863;Unigene2712810;Unigene714021;Unigene3845994;Unigene3131592;Unigene2518195;Unigene2762224;Unigene1022367;Unigene1751303;Unigene3349790;Unigene162163;Unigene3772048;Unigene1251860;Unigene504918;Unigene401732;Unigene3844653;Unigene3698682;Unigene3823752;Unigene2133776;Unigene1504541;Unigene887552;Unigene4676684;Unigene3493152;Unigene2639631;Unigene804994;Unigene1094811;Unigene424265;Unigene2236702;Unigene750014;Unigene1032916;Unigene2681268;Unigene4320963;Unigene1610154;Unigene162102;Unigene251582;Unigene941689;Unigene1389636;Unigene1311935;Unigene2011318;Unigene3331311;Unigene362320;Unigene4368588;Unigene4861482;Unigene3131676;Unigene142038;Unigene1795414;Unigene493342;Unigene1861681;Unigene3312625;Unigene2198750;Unigene3273130;Unigene1978770;Unigene924277;Unigene2746313;Unigene815311;Unigene830616;Unigene1383440;Unigene4481957;Unigene2893464;Unigene3938085;Unigene1269755;Unigene1640174;Unigene1348473;Unigene1447911;Unigene3340297;Unigene1641397;Unigene2617896;Unigene4689720;Unigene1684416;Unigene3885101;Unigene4823704;Unigene3375387;Unigene4328461;Unigene4169828;Unigene67021;Unigene4256362;Unigene3421763;Unigene223354;Unigene1830226;Unigene3533193;Unigene3301845;Unigene1211877;Unigene2548734;Unigene3303257;Unigene367537;Unigene1597642;Unigene950846;Unigene1676102;Unigene1513841;Unigene3708624;Unigene4378966;Unigene1059123;Unigene4356255;Unigene1586407;Unigene2359567;Unigene1951572;Unigene3044161;Unigene2250146;Unigene95412;Unigene3658059;Unigene1228748;Unigene4379429;Unigene1645738;Unigene1885497;Unigene1769428;Unigene714415;Unigene4480579;Unigene1468504;Unigene2830394;Unigene622191;Unigene1917940;Unigene888704;Unigene1438882;Unigene2014981;Unigene36176;Unigene236559;Unigene3671203;Unigene3950955;Unigene3664290;Unigene2059338;Unigene3470916;Unigene2549547;Unigene2330343;Unigene2855193;Unigene388881;Unigene854069;Unigene2307910;Unigene84699;Unigene4429499;Unigene2029958;Unigene579229;Unigene2684387;Unigene2178587;Unigene3643856;Unigene1668409;Unigene1337681;Unigene1143183;Unigene1766290;Unigene44986;Unigene4898273;Unigene1924218;Unigene4615570;Unigene2767628;Unigene1391220;Unigene1902462;Unigene4033752;Unigene3892259;Unigene2390741;Unigene406758;Unigene4366642;Unigene2848650;Unigene2552545;Unigene649892;Unigene4020926;Unigene3121405;Unigene2440875;Unigene3545278;Unigene1460915;Unigene220098;Unigene4627472;Unigene4623163;Unigene9928;Unigene4720271;Unigene468346;Unigene351273;Unigene4381241;Unigene3042600;Unigene1281628;Unigene257619;Unigene4655016;Unigene1249286;Unigene1400459;Unigene3472994;Unigene4861967;Unigene2470552;Unigene2336487;Unigene1743558;Unigene3485227;Unigene1162792;Unigene3713726;Unigene4184712;Unigene3807377;Unigene4250812;Unigene1032209;Unigene2077515;Unigene1004853;Unigene409637;Unigene1291803;Unigene1814817;Unigene1417356;Unigene2577855;Unigene969632;Unigene2470553;Unigene3132922;Unigene78553;Unigene4648040;Unigene1611681;Unigene1803224;Unigene4521959;Unigene1499675;Unigene3281435;Unigene2364774;Unigene1927204;Unigene3271890;Unigene1821620;Unigene211492;Unigene557363;Unigene625100;Unigene492167;Unigene1216422;Unigene4609894;Unigene1636045;Unigene267294;Unigene893019;Unigene1398429;Unigene2287559;Unigene1036964;Unigene824976;Unigene1138136;Unigene1732625;Unigene3157136;Unigene839119;Unigene482126;Unigene1686774;Unigene1122936;Unigene1107978;Unigene1952314;Unigene2411400;Unigene4793831;Unigene493691;Unigene2115978;Unigene3430496;Unigene2032201;Unigene4851219;Unigene1567800;Unigene2864782;Unigene2946008;Unigene357362;Unigene3414649;Unigene3480995;Unigene4824505;Unigene4306201;Unigene4536023;Unigene4347616;Unigene4886766;Unigene443213;Unigene1076343;Unigene1132352;Unigene2733460;Unigene997751;Unigene1161707;Unigene430867;Unigene4701663;Unigene783745;Unigene3253741;Unigene133197;Unigene904536;Unigene3259877;Unigene242529;Unigene3922870;Unigene2392676;Unigene612457;Unigene1291802;Unigene2756384;Unigene1640623;Unigene2734935;Unigene1661982;Unigene626345;Unigene4745282;Unigene434563;Unigene4843078;Unigene3467891;Unigene4523805;Unigene1473355;Unigene3751955;Unigene159742;Unigene221011;Unigene3777474;Unigene2299460;Unigene2910618;Unigene2771419;Unigene742441;Unigene3423909;Unigene1624762;Unigene2670060;Unigene2181951;Unigene2053668;Unigene3883520;Unigene4533354;Unigene1552206;Unigene823040;Unigene3527005;Unigene3959054;Unigene1861876;Unigene792235;Unigene2667938;Unigene428944;Unigene954283;Unigene991585;Unigene2893556;Unigene532228;Unigene2634557;Unigene1200177;Unigene1481250;Unigene2610286;Unigene4144897;Unigene894786;Unigene1395655;Unigene72699;Unigene4580359;Unigene3640040;Unigene3462357;Unigene4134628;Unigene2572142;Unigene1774260;Unigene3415882;Unigene1303318;Unigene177756;Unigene2525807;Unigene1680243;Unigene2753570;Unigene1097573;Unigene432307;Unigene2183146;Unigene4484146;Unigene3524616;Unigene1133843;Unigene2505068;Unigene1995802;Unigene472007;Unigene2561519;Unigene814363;Unigene1220869;Unigene85279;Unigene694436;Unigene671037;Unigene4057058;Unigene2289363;Unigene619357;Unigene1602887;Unigene114478;Unigene2398323;Unigene1734389;Unigene2454448;Unigene1103047;Unigene243700;Unigene958863;Unigene1135706;Unigene4433188;Unigene1205610;Unigene4183763;Unigene4743316;Unigene3208088;Unigene1474856;Unigene1078041;Unigene252882;Unigene764206;Unigene3922528;Unigene2301184;Unigene980074;Unigene1000715;Unigene4137455;Unigene3998619;Unigene3150753;Unigene602755;Unigene2424176;Unigene173692;Unigene1516304;Unigene450647;Unigene3185093;Unigene3848465;Unigene505368;Unigene3879214;Unigene1625690;Unigene3461210;Unigene3615446;Unigene2996198;Unigene3978482;Unigene3231464;Unigene521236;Unigene4284940;Unigene4384366;Unigene3389736;Unigene1676089;Unigene2756655;Unigene659105;Unigene788130;Unigene1953827;Unigene3589907;Unigene4468484;Unigene103929;Unigene510551;Unigene3171832;Unigene1120605;Unigene932410;Unigene353062;Unigene742847;Unigene154442;Unigene4218582;Unigene2488976;Unigene81333;Unigene1081146;Unigene3669187;Unigene4818030;Unigene4844723;Unigene4837529;Unigene276586;Unigene299880;Unigene2898531;Unigene2652495;Unigene2752868;Unigene4506942;Unigene2360847;Unigene4192350;Unigene289661;Unigene2186638;Unigene2779521;Unigene3130928;Unigene28790;Unigene2826241;Unigene3505727;Unigene4700448;Unigene26062;Unigene4102988;Unigene4696535;Unigene315006;Unigene4263476;Unigene1454960;Unigene4490095;Unigene4158115;Unigene506021;Unigene376399;Unigene3848532;Unigene674661;Unigene2653475;Unigene394425;Unigene4011097;Unigene959319;Unigene1366988;Unigene4757884;Unigene3882932;Unigene117447;Unigene1111635;Unigene2009138;Unigene4348144;Unigene3803817;Unigene4342015;Unigene380430;Unigene1485837;Unigene2523260;Unigene1196218;Unigene2944468;Unigene4332572;Unigene4121824;Unigene3738883;Unigene4417577;Unigene3143542;Unigene3997978;Unigene3795173;Unigene1506349;Unigene3623877;Unigene2698506;Unigene4643203;Unigene485323;Unigene3021038;Unigene796182;Unigene3500573;Unigene3900123;Unigene3233215;Unigene2908644;Unigene3006548;Unigene1531932;Unigene1644104;Unigene913197;Unigene4467786;Unigene4904683;Unigene858561;Unigene506665;Unigene2263568;Unigene4469343;Unigene993397;Unigene4758314;Unigene1943360;Unigene4347971;Unigene1297572;Unigene2684582;Unigene1706547;Unigene3008649;Unigene4403116;Unigene3272279;Unigene3900863;Unigene4590123;Unigene4537333;Unigene1794759;Unigene2069532;Unigene3794865;Unigene470486;Unigene2241363;Unigene1762211;Unigene3656808;Unigene237499;Unigene2533705;Unigene1964209;Unigene4635504;Unigene1838122;Unigene933479;Unigene4026824;Unigene1921717;Unigene4270788;Unigene3562912;Unigene3834456;Unigene4083881;Unigene1420788;Unigene2889103;Unigene3453685;Unigene4812904;Unigene2689995;Unigene2798430;Unigene855685;Unigene4410995;Unigene2306640;Unigene3202544;Unigene2949331;Unigene3679969;Unigene1196640;Unigene1951790;Unigene1369892;Unigene985776;Unigene1495374;Unigene1071446;Unigene3163993;Unigene2560605;Unigene1828430;Unigene3591832;Unigene4453492;Unigene4487105;Unigene1265163;Unigene1944269;Unigene1601226;Unigene118838;Unigene2870917;Unigene4306948;Unigene1872986;Unigene4643033;Unigene3138068;Unigene3391380;Unigene4416298;Unigene3831819;Unigene2077161;Unigene2867292;Unigene354329;Unigene1912371;Unigene1562784;Unigene957501;Unigene1406289;Unigene236532;Unigene1665446;Unigene3023616;Unigene4832474;Unigene3373534;Unigene4917815;Unigene2606464;Unigene1735465;Unigene755514;Unigene2730035;Unigene1997630;Unigene4164226;Unigene2159425;Unigene3142241;Unigene2204989;Unigene3762122;Unigene1933787;Unigene1760563;Unigene1241122;Unigene2756841;Unigene1932472;Unigene4370423;Unigene3956931;Unigene4193375;Unigene2558995;Unigene1295699;Unigene4823938;Unigene1988259;Unigene394704;Unigene245346;Unigene1397229;Unigene3290473;Unigene393341;Unigene3651108;Unigene4479950;Unigene3238796;Unigene3375853;Unigene4742540;Unigene4363995;Unigene3887256;Unigene2320063;Unigene1642051;Unigene3021783;Unigene1304130;Unigene1816097;Unigene3055609;Unigene965487;Unigene2124585;Unigene4781326;Unigene460704;Unigene1319446;Unigene1739774;Unigene3414743;Unigene4461156;Unigene3973590;Unigene4509001;Unigene1753881;Unigene1541041;Unigene1804432;Unigene3789661;Unigene2205130;Unigene3027129;Unigene2561644;Unigene326210;Unigene4159681;Unigene1325556;Unigene3426177;Unigene2799194;Unigene2605188;Unigene4706387;Unigene3126271;Unigene1240176;Unigene498721;Unigene1000037;Unigene1532983;Unigene2187991;Unigene3670099;Unigene585692;Unigene4019152;Unigene1792533;Unigene3139498;Unigene1196991;Unigene3697023;Unigene1243412;Unigene3611183;Unigene1324361;Unigene4468809;Unigene4943921;Unigene4806898;Unigene52813;Unigene2583282;Unigene789163;Unigene311705;Unigene373869;Unigene3318063;Unigene648605;Unigene2269582;Unigene376077;Unigene1799285;Unigene180810;Unigene1355424;Unigene865382;Unigene3869515;Unigene510043;Unigene4758833;Unigene4077249;Unigene1238547;Unigene3836083;Unigene3037590;Unigene1761782;Unigene246165;Unigene3268861;Unigene4775018;Unigene1273786;Unigene2818571;Unigene1587589;Unigene27750;Unigene2759046;Unigene18588;Unigene3813484;Unigene327065;Unigene1572419;Unigene784969;Unigene3913258;Unigene1575444;Unigene3792336;Unigene4270373;Unigene463139;Unigene1260077;Unigene4530146;Unigene4112165;Unigene1546747;Unigene86150;Unigene4415453;Unigene2949035;Unigene4893967;Unigene637492;Unigene659586;Unigene888790;Unigene3829296;Unigene3522895;Unigene4127492;Unigene579169;Unigene806259;Unigene1668878;Unigene4934563;Unigene3004474;Unigene4810389;Unigene2201795;Unigene3336406;Unigene1484402;Unigene4751701;Unigene1539622;Unigene1919262;Unigene4415374;Unigene2931575;Unigene1734356;Unigene2486371;Unigene2810931;Unigene107613;Unigene387076;Unigene412694;Unigene844318;Unigene655727;Unigene2996053;Unigene3821079;Unigene2833185;Unigene3369962;Unigene2604020;Unigene3916848;Unigene3367702;Unigene827457;Unigene769598;Unigene948938;Unigene3003013;Unigene4533077;Unigene2220181;Unigene2095248;Unigene2076597;Unigene4135531;Unigene1662533;Unigene3568909;Unigene1553604;Unigene3267167;Unigene3976050;Unigene2912920;Unigene3650663;Unigene829191;Unigene3836272;Unigene1863966;Unigene688359;Unigene2905456;Unigene429963;Unigene1331114;Unigene1824218;Unigene1525803;Unigene2004769;Unigene2187662;Unigene884394;Unigene899820;Unigene4681465;Unigene2928774;Unigene2749112;Unigene2558932;Unigene2024628;Unigene3063997;Unigene1508534;Unigene2508769;Unigene4790984;Unigene790443;Unigene3021263;Unigene4440397;Unigene414112;Unigene737669;Unigene1625446;Unigene1125300;Unigene3501089;Unigene4657432;Unigene1876724;Unigene2944464;Unigene3050651;Unigene330711;Unigene204447;Unigene4130576;Unigene4012364;Unigene3212835;Unigene1132148;Unigene2261892;Unigene2956252;Unigene3151777;Unigene3898447;Unigene793388;Unigene120353;Unigene584508;Unigene1912322;Unigene2443422;Unigene1516135;Unigene2425724;Unigene1848939;Unigene1941900;Unigene167363;Unigene4354521;Unigene2080089;Unigene4909959;Unigene4173838;Unigene2547228;Unigene2594597;Unigene4128688;Unigene2287192;Unigene2364111;Unigene126415;Unigene485821;Unigene3823109;Unigene1633994;Unigene2760748;Unigene422493;Unigene504018;Unigene2150907;Unigene3833570;Unigene2940897;Unigene2212613;Unigene1618498;Unigene2864619;Unigene2081004;Unigene1796300;Unigene3879515;Unigene4586273;Unigene4405848;Unigene205129;Unigene447409;Unigene1981395;Unigene221522;Unigene3968386;Unigene2092197;Unigene1966356;Unigene1374533;Unigene3174798;Unigene2848100;Unigene428513;Unigene973633;Unigene4004811;Unigene1408670;Unigene2389797;Unigene788157;Unigene1793317;Unigene3689250;Unigene583071;Unigene250181;Unigene1404700;Unigene72121;Unigene2394133;Unigene336114;Unigene3367151;Unigene3150305;Unigene1413638;Unigene287166;Unigene2388991;Unigene997142;Unigene3535736;Unigene4013531;Unigene3800374;Unigene1494181;Unigene780787;Unigene1271708;Unigene4031264;Unigene2981866;Unigene910829;Unigene581746;Unigene2920935;Unigene2657944;Unigene254563;Unigene3407910;Unigene2528951;Unigene623282;Unigene4110642;Unigene4876763;Unigene2871106;Unigene1596799;Unigene4911220;Unigene1789444;Unigene4090377;Unigene3940573;Unigene839049;Unigene1494115;Unigene1861879;Unigene314108;Unigene1671599;Unigene345355;Unigene837317;Unigene2062465;Unigene2801521;Unigene4363218;Unigene3351866;Unigene2960553;Unigene253467;Unigene563904;Unigene2057916;Unigene3722730;Unigene1228356;Unigene669631;Unigene1136327;Unigene3018346;Unigene3382991;Unigene363806;Unigene3415331;Unigene454802;Unigene51950;Unigene1949115;Unigene2081633;Unigene1804221;Unigene3891922;Unigene429830;Unigene111165;Unigene1773932;Unigene711659;Unigene415847;Unigene3138044;Unigene69658;Unigene2023211;Unigene3835438;Unigene2975542;Unigene2067403;Unigene566867;Unigene1506273;Unigene4686153;Unigene247539;Unigene4242775;Unigene1285650;Unigene4432875;Unigene2184960;Unigene1178268;Unigene2174265;Unigene2845083;Unigene2819996;Unigene2473041;Unigene3716331;Unigene4118631;Unigene3664697;Unigene1034570;Unigene4348319;Unigene4312214;Unigene1383444;Unigene4190220;Unigene2845148;Unigene671929;Unigene4028516;Unigene1538378;Unigene3859139;Unigene555704;Unigene3678244;Unigene91882;Unigene1207560;Unigene1226936;Unigene1903992;Unigene1657715;Unigene4166393;Unigene3227396;Unigene3477931;Unigene619544;Unigene2178090;Unigene4312242;Unigene1129612;Unigene3721642;Unigene2544650;Unigene283282;Unigene915421;Unigene2007586;Unigene202684;Unigene3341580;Unigene4789770;Unigene3131865;Unigene1297666;Unigene2351173;Unigene853527;Unigene1366460;Unigene2069227;Unigene222196;Unigene4395525;Unigene4311697;Unigene62968;Unigene4231180;Unigene2981470;Unigene2170890;Unigene883917;Unigene887841;Unigene1463700;Unigene1791573;Unigene2099664;Unigene3701193;Unigene4636747;Unigene2142867;Unigene2013625;Unigene2874394;Unigene3364564;Unigene4738867;Unigene6885;Unigene3110948;Unigene3870923;Unigene1042836;Unigene3688351;Unigene3523975;Unigene3658858;Unigene1445454;Unigene279069;Unigene2339472;Unigene4429631;Unigene4293687;Unigene1903378;Unigene1228930;Unigene4261481;Unigene2901605;Unigene2298298;Unigene4785059;Unigene4198945;Unigene1869423;Unigene4112893;Unigene2609055;Unigene3793358;Unigene3628561;Unigene2203859;Unigene1938000;Unigene3360716;Unigene1842588;Unigene3457925;Unigene1849653;Unigene860726;Unigene876431;Unigene2540178;Unigene570460;Unigene3836337;Unigene3807730;Unigene3642119;Unigene1529760;Unigene3522997;Unigene4395311;Unigene198023;Unigene47001;Unigene342940;Unigene4944210;Unigene3319182;Unigene1643296;Unigene3143463;Unigene2738315;Unigene1906926;Unigene1439313;Unigene708919;Unigene2454376;Unigene4864156;Unigene2523372;Unigene4318508;Unigene2710439;Unigene242280;Unigene4315984;Unigene2346839;Unigene1464226;Unigene474414;Unigene4112712;Unigene1616068;Unigene556107;Unigene4208557;Unigene2866424;Unigene4058637;Unigene2897332;Unigene4013267;Unigene4951424;Unigene1003852;Unigene3506056;Unigene854188;Unigene3094156;Unigene950694;Unigene3270791;Unigene1845879;Unigene1573923;Unigene4416;Unigene3275922;Unigene281042;Unigene504626;Unigene2547916;Unigene4474511;Unigene29874;Unigene3085250;Unigene201211;Unigene4005307;Unigene4102962;Unigene384078;Unigene3517621;Unigene4928205;Unigene2793154;Unigene497624;Unigene1829840;Unigene577032;Unigene727327;Unigene3840135;Unigene2703506;Unigene1427567;Unigene958330;Unigene553988;Unigene4345302;Unigene512589;Unigene4830231;Unigene277008;Unigene2101138;Unigene3121571;Unigene4514466;Unigene4307560;Unigene865751;Unigene3985416;Unigene49029;Unigene426667;Unigene4354941;Unigene3005265;Unigene338312;Unigene4304153;Unigene23176;Unigene3914984;Unigene2735015;Unigene4948848;Unigene2111492;Unigene2802200;Unigene2170222;Unigene393943;Unigene558060;Unigene2070108;Unigene3450105;Unigene1454807;Unigene2042078;Unigene3742609;Unigene2643253;Unigene878239;Unigene4285699;Unigene267635;Unigene838685;Unigene401530;Unigene657391 |
| AA | Auxiliary Activities | AA10 | AA10 (formerly CBM33) proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs); some proteins have been shown to act on chitin, others on cellulose; lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54); lytic cellulose monooxygenase (C4-dehydrogenating)(EC 1.14.99.56); lytic chitin monooxygenase (EC 1.14.99.53) | 80 | Unigene1336444;Unigene2779114;Unigene615694;Unigene4789776;Unigene3971533;Unigene1428158;Unigene3127938;Unigene1619422;Unigene210738;Unigene4365491;Unigene114648;Unigene2538497;Unigene4202712;Unigene2183136;Unigene3789908;Unigene487979;Unigene547741;Unigene219971;Unigene1061865;Unigene4756387;Unigene470419;Unigene3702929;Unigene2253288;Unigene2859609;Unigene1244518;Unigene3453626;Unigene3521786;Unigene729878;Unigene3225703;Unigene4356596;Unigene4830395;Unigene4145028;Unigene2259250;Unigene973514;Unigene4710403;Unigene1431077;Unigene719247;Unigene283936;Unigene1412707;Unigene1531288;Unigene3996556;Unigene3218378;Unigene4083558;Unigene1342273;Unigene112211;Unigene1319088;Unigene3670255;Unigene1645165;Unigene3831428;Unigene3174080;Unigene3812608;Unigene2025798;Unigene1924441;Unigene3534355;Unigene4546245;Unigene2659199;Unigene3359250;Unigene2986791;Unigene276214;Unigene1301416;Unigene4830396;Unigene1882924;Unigene991308;Unigene4106892;Unigene56176;Unigene3446134;Unigene4555996;Unigene1556710;Unigene2785304;Unigene3759124;Unigene2986728;Unigene817611;Unigene1140905;Unigene4305080;Unigene3037987;Unigene714729;Unigene352088;Unigene3527243;Unigene3326466;Unigene411142 |
| AA | Auxiliary Activities | AA3 | cellobiose dehydrogenase (EC 1.1.99.18); glucose 1-oxidase (EC 1.1.3.4); aryl alcohol oxidase (EC 1.1.3.7); alcohol oxidase (EC 1.1.3.13); pyranose oxidase (EC 1.1.3.10) | 14 | Unigene3887080;Unigene4031013;Unigene614775;Unigene3367347;Unigene1545151;Unigene16187;Unigene113985;Unigene3295999;Unigene3150810;Unigene3364816;Unigene2140525;Unigene253144;Unigene4770530;Unigene3693298 |
| AA | Auxiliary Activities | AA4 | vanillyl-alcohol oxidase (EC 1.1.3.38) | 132 | Unigene1657095;Unigene1984421;Unigene1322280;Unigene657828;Unigene461217;Unigene2732914;Unigene2195235;Unigene1556358;Unigene79372;Unigene2410796;Unigene4278904;Unigene1481962;Unigene175410;Unigene4879639;Unigene110715;Unigene190021;Unigene2110170;Unigene726874;Unigene2020114;Unigene4052204;Unigene1000261;Unigene1240705;Unigene2732911;Unigene2319642;Unigene1526785;Unigene2000594;Unigene379656;Unigene857619;Unigene957920;Unigene4618211;Unigene499107;Unigene717069;Unigene225700;Unigene133697;Unigene1603421;Unigene1945118;Unigene399925;Unigene3694344;Unigene556231;Unigene1230816;Unigene1406920;Unigene450036;Unigene1406919;Unigene1913662;Unigene3874950;Unigene4385032;Unigene1756537;Unigene1084029;Unigene4438520;Unigene2206424;Unigene4625490;Unigene1943455;Unigene1934765;Unigene3004728;Unigene1871934;Unigene1793535;Unigene304691;Unigene145441;Unigene529818;Unigene1077196;Unigene3591195;Unigene4168305;Unigene555198;Unigene2023782;Unigene910630;Unigene3496129;Unigene2576373;Unigene152662;Unigene4627776;Unigene147356;Unigene430294;Unigene4879058;Unigene40465;Unigene4783104;Unigene1018801;Unigene3308616;Unigene918060;Unigene82972;Unigene2597394;Unigene4785801;Unigene712417;Unigene880324;Unigene2229020;Unigene163512;Unigene2166383;Unigene3143947;Unigene2536265;Unigene2322099;Unigene2714573;Unigene4346401;Unigene850791;Unigene3308618;Unigene4440799;Unigene4470421;Unigene258530;Unigene4219798;Unigene1735067;Unigene122884;Unigene2400788;Unigene4889315;Unigene2732913;Unigene2173423;Unigene2884478;Unigene866729;Unigene1140060;Unigene2624535;Unigene3496130;Unigene3963384;Unigene1210375;Unigene699691;Unigene1350254;Unigene190019;Unigene1894368;Unigene853204;Unigene1589628;Unigene1140439;Unigene632994;Unigene1459196;Unigene898298;Unigene331196;Unigene811611;Unigene4271816;Unigene943547;Unigene2273879;Unigene4107084;Unigene2195243;Unigene4440797;Unigene1630271;Unigene518231;Unigene290530;Unigene177274;Unigene2365540 |
| AA | Auxiliary Activities | AA5 | Oxidase with oxygen as acceptor (EC 1.1.3.-); galactose oxidase (EC 1.1.3.9); glyoxal oxidase (EC 1.2.3.15); alcohol oxidase (EC 1.1.3.13) | 118 | Unigene1319503;Unigene908048;Unigene144854;Unigene4452840;Unigene455511;Unigene1305525;Unigene2554893;Unigene1155881;Unigene722667;Unigene1657919;Unigene1377059;Unigene31302;Unigene2883062;Unigene4594998;Unigene2242292;Unigene164000;Unigene279800;Unigene3621983;Unigene4060887;Unigene4181118;Unigene1935169;Unigene4092916;Unigene638549;Unigene4924052;Unigene2583032;Unigene2728443;Unigene1833627;Unigene4384257;Unigene1468652;Unigene2568943;Unigene628989;Unigene3433055;Unigene3869597;Unigene1905175;Unigene1442848;Unigene836127;Unigene2897736;Unigene668966;Unigene51348;Unigene2804513;Unigene41203;Unigene67006;Unigene2309433;Unigene1772253;Unigene1695644;Unigene132987;Unigene1225370;Unigene3178383;Unigene1975840;Unigene809052;Unigene2476453;Unigene657085;Unigene576994;Unigene768466;Unigene1120178;Unigene2740603;Unigene1124894;Unigene2232523;Unigene799301;Unigene1581420;Unigene4533775;Unigene822461;Unigene3101130;Unigene1665450;Unigene2627624;Unigene1724450;Unigene263074;Unigene918901;Unigene4680561;Unigene8314;Unigene1129241;Unigene530183;Unigene339947;Unigene2848975;Unigene524103;Unigene4502841;Unigene3025468;Unigene256250;Unigene2404723;Unigene1121810;Unigene946136;Unigene1500512;Unigene1503664;Unigene4746890;Unigene1632698;Unigene1124343;Unigene2416854;Unigene540238;Unigene2756887;Unigene772242;Unigene3725579;Unigene954009;Unigene652287;Unigene2069311;Unigene3280879;Unigene616941;Unigene329471;Unigene1167525;Unigene1115709;Unigene290940;Unigene1621559;Unigene327457;Unigene1241855;Unigene92789;Unigene1685388;Unigene1292986;Unigene2914626;Unigene2465691;Unigene1467423;Unigene3495275;Unigene1110342;Unigene547451;Unigene1486382;Unigene2612236;Unigene1759878;Unigene2235522;Unigene189029;Unigene2253928 |
| AA | Auxiliary Activities | AA6 | 1,4-benzoquinone reductase (EC. 1.6.5.6) | 4 | Unigene3560931;Unigene304629;Unigene1271565;Unigene615123 |
| AA | Auxiliary Activities | AA7 | glucooligosaccharide oxidase (EC 1.1.3.-); chitooligosaccharide oxidase (EC 1.1.3.-) | 11 | Unigene2001300;Unigene3297112;Unigene12290;Unigene742898;Unigene2695095;Unigene2886105;Unigene110715;Unigene251846;Unigene3416721;Unigene285326;Unigene843749 |
| AA | Auxiliary Activities | AA8 | Iron reductase domain | 1 | Unigene253144 |
| AA | Auxiliary Activities | AA9 | AA9 (formerly GH61) proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs); cleavage of cellulose chains with oxidation of carbons C1 and/or C4 and C-6); lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54); lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56) | 3 | Unigene74575;Unigene3577271;Unigene1041337 |
使用柱形图展示每个功能分类的基因数目,便于对比微生物群体中各功能分类的基因数量分布情况。
![]() |
Fig 6-3-1 CAZy注释统计图 |
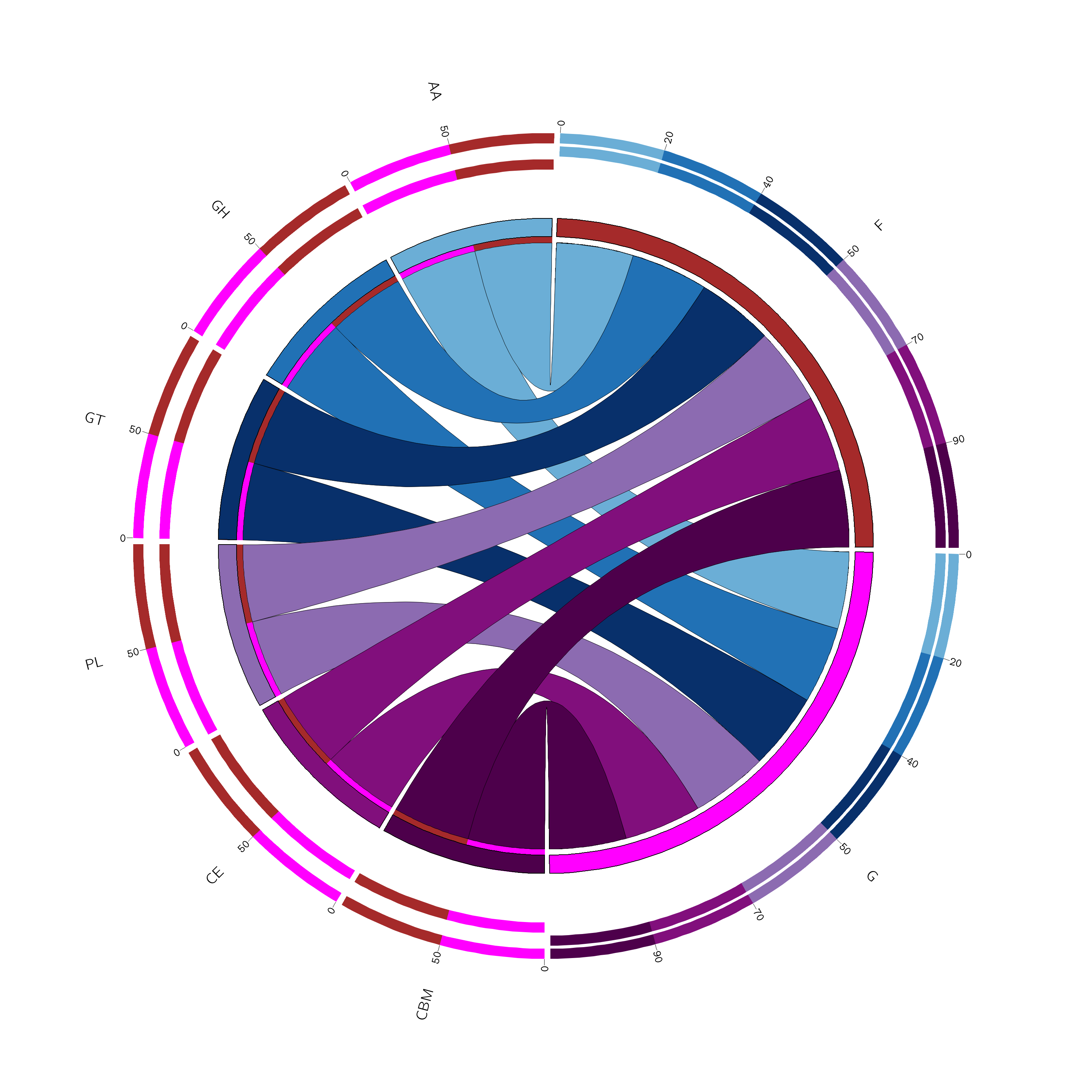
我们使用circos软件绘制circos图展示每个功能分类的丰度分布,便于了解各样本功能分布规律。
![]() |
Fig 6-3-2 CAZy功能分布circos图 |
6.4 CARD注释
微生物抗性及毒性基因的研究,是微生物研究中极具应用价值和实际应用意义的一项研究,目前我们常用的方法大多是依赖相关抗性毒性基因数据库进行分析。
CARD(Comprehensive Antibiotic Resistance Database)以Antibiotic Resistance Ontology(ARO)为分类单位(term)构建,用于关联抗生素模块及其目标、抗性机制、基因变异等信息。通过该数据库的注释,可以找到耐药性相关基因的名称,所耐受的抗生素种类等信息。
我们使用 Diamond 将预测基因序列比对至数据库进行注释
Tab 6-4-1 CARD注释表
| GeneID | ARO_Name | ARO_Accession | AMR_Gene_Family | Drug_Class | Resistance_Mechanism | F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 |
|---|
| Unigene60 | YojI | ARO:3003952 | ATP-binding cassette (ABC) antibiotic efflux pump | peptide antibiotic | antibiotic efflux | 3.556833891525611e-07 | 3.192848354869383e-07 | 3.85283918966469e-07 | 0 | 0 | 0 | 1.222359715024187e-07 | 0 |
| Unigene148 | TaeA | ARO:3003986 | ATP-binding cassette (ABC) antibiotic efflux pump | pleuromutilin antibiotic | antibiotic efflux | 0 | 5.455598110104968e-07 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene219 | tetA(58) | ARO:3003980 | major facilitator superfamily (MFS) antibiotic efflux pump | tetracycline antibiotic | antibiotic efflux | 0 | 1.7589275704065382e-06 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene413 | vanSF | ARO:3002936 | glycopeptide resistance gene cluster;vanS | glycopeptide antibiotic | antibiotic target alteration | 7.479896509593455e-06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene414 | vanRF | ARO:3002925 | glycopeptide resistance gene cluster;vanR | glycopeptide antibiotic | antibiotic target alteration | 6.132217069531436e-06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene433 | Streptomyces rishiriensis parY mutant conferring resistance to aminocoumarin | ARO:3003318 | aminocoumarin resistant parY | aminocoumarin antibiotic | antibiotic target alteration | 1.7962707146585014e-07 | 8.522954737313404e-07 | 0 | 0 | 0 | 0 | 0 | 3.46766079062987e-06 |
| Unigene483 | vanG | ARO:3002909 | glycopeptide resistance gene cluster;van ligase | glycopeptide antibiotic | antibiotic target alteration | 1.7835037269058013e-06 | 0 | 0 | 0 | 4.1285748068600216e-07 | 0 | 0 | 0 |
| Unigene712 | vanSE | ARO:3002935 | glycopeptide resistance gene cluster;vanS | glycopeptide antibiotic | antibiotic target alteration | 6.488473254415475e-06 | 5.296696246067359e-06 | 6.960216761679971e-06 | 3.0931989253593805e-06 | 0 | 5.255303825025871e-07 | 3.247371673999093e-07 | 0 |
| Unigene792 | bcrA | ARO:3002987 | ATP-binding cassette (ABC) antibiotic efflux pump | peptide antibiotic | antibiotic efflux | 1.7200524161226133e-06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene793 | bcrA | ARO:3002987 | ATP-binding cassette (ABC) antibiotic efflux pump | peptide antibiotic | antibiotic efflux | 2.992257083346942e-06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
我们使用circos软件绘制circos图展现丰度排名前10的ARO功能分布规律,便于初步比较样本间抗性基因功能分布差异等信息。
![]() |
Fig 6-4-1 CARD注释Circos图 |
6.5 VFDB注释
VFDB,Virulence Factors of Pathogenic Bacteria,毒力因子数据库,由中国医学科学院研发,收集整理了重要医学病原菌中74个属951个菌株的1080个已知毒力因子的组成、结构、功能、致病机理、毒力岛、序列和基因组信息等内容。毒力因子数据库能够为病原菌的入侵机理研究提供关键证据,可以研究群落中不同病原菌的毒力因子构成等特征,被广泛应用于毒力因子基因鉴定。
我们使用 Diamond 与数据库进行比对注释
Tab 6-5-1 VFDB注释表
| GeneID | Subject | Description | Organism | VFs | VF_FullName | Function | VFID | F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 |
|---|
| Unigene1 | VFG022639(gi:333989135) | (proC) pyrroline-5-carboxylate reductase ProC [Proline synthesis (CVF307)] [Mycobacterium sp. JDM601] | - | - | - | - | - | 9.038851272516904e-07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene16 | VFG031407(gi:433630073) | (ctpV) Putative metal cation transporter P-type ATPase CtpV [Copper exporter (CVF658)] [Mycobacterium canettii CIPT 140070010] | - | - | - | - | - | 0 | 0 | 1.8493628110390513e-07 | 2.4037713864265717e-07 | 0 | 2.967263527620739e-06 | 0 | 0 |
| Unigene60 | VFG044172(gb|NP_756180) | (chuV) ATP-binding hydrophilic protein ChuV [Chu (VF0227)] [Escherichia coli CFT073] | Escherichia coli (UPEC) | Chu | E. coli hemin uptake | Iron uptake: the ability to use heme and/or hemoglobin might be especially advantageous to pathogenic bacteria. These pathogens often secrete cytotoxins, which gain access to the intracellular heme reservoir besides initiating tissue invasion. Cytotoxin production coupled with the capability to utilize heme and/or hemoglobin could serve as an effective iron acquisition strategy during the progression of infection | VF0227 | 3.556833891525611e-07 | 3.192848354869383e-07 | 3.85283918966469e-07 | 0 | 0 | 0 | 1.222359715024187e-07 | 0 |
| Unigene144 | VFG031084(gi:433644988) | (zmp1) endothelin-converting enzyme [Zn++ metallophrotease (CVF655)] [Mycobacterium smegmatis JS623] | - | - | - | - | - | 2.4816795455809075e-05 | 2.1444175455707123e-05 | 1.8403891096352753e-05 | 1.0683850988088986e-05 | 3.628834343687883e-05 | 6.83472219294583e-05 | 9.362398618977449e-05 | 2.465023983293573e-06 |
| Unigene148 | VFG007913(gi:145223975) | (ddrA) daunorubicin resistance ABC transporter ATPase subunit [PDIM (phthiocerol dimycocerosate) and PGL (phenolic glycolipid) biosynthesis and transport (CVF288)] [Mycobacterium gilvum PYR-GCK] | - | - | - | - | - | 0 | 5.455598110104968e-07 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene171 | VFG014970(gi:148545464) | (algR) two component transcriptional regulator, LytTR family [Alginate regulation (CVF523)] [Pseudomonas putida F1] | - | - | - | - | - | 0 | 2.6159762366466053e-07 | 0 | 3.461953355451312e-07 | 0 | 0 | 0 | 0 |
| Unigene211 | VFG009927(gi:118617353) | (relA) GTP pyrophosphokinase RelA [(p)ppGpp synthesis and hydrolysis (CVF335)] [Mycobacterium ulcerans Agy99] | - | - | - | - | - | 0 | 1.8113238601911703e-06 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene219 | VFG044173(gi:28901507) | (pvuE) iron-dicitrate transporter ATP-binding subunit [vibrioferrin (IA038)] [Vibrio parahaemolyticus RIMD 2210633] | - | - | - | - | - | 0 | 1.7589275704065382e-06 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene236 | VFG037640(gi:169794956) | (bap) hypothetical protein [Biofilm-associated protein (CVF771)] [Acinetobacter baumannii AYE] | - | - | - | - | - | 5.098870247118811e-06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene253 | VFG016390(gi:42784429) | (BCE_5384) NAD dependent epimerase/dehydratase family protein [Polysaccharide capsule (CVF567)] [Bacillus cereus ATCC 10987] | - | - | - | - | - | 1.4613348546578092e-07 | 2.6235808187298805e-07 | 0 | 0 | 0 | 0 | 0 | 0 |
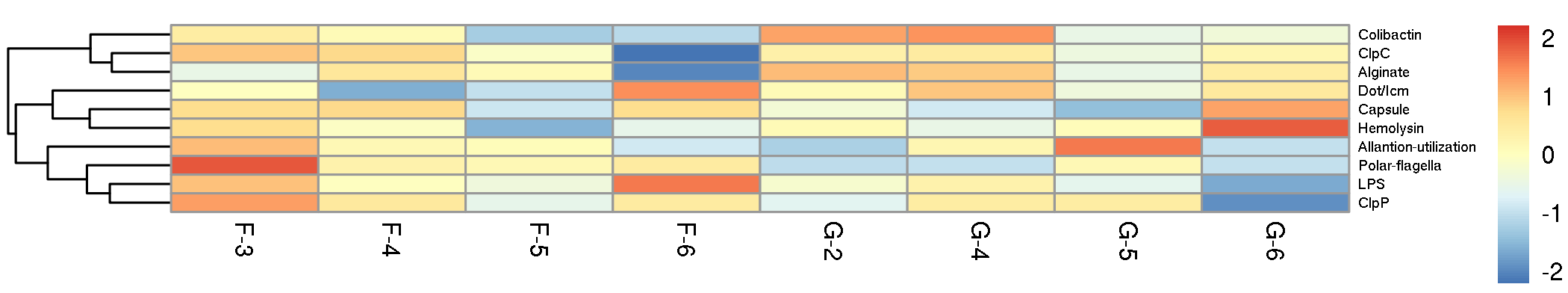
我们挑选丰度排名前10的毒力因子,以热图展示其在各样本间的分布规律:
![]() |
Fig 6-5-1 毒力因子丰度热图 |
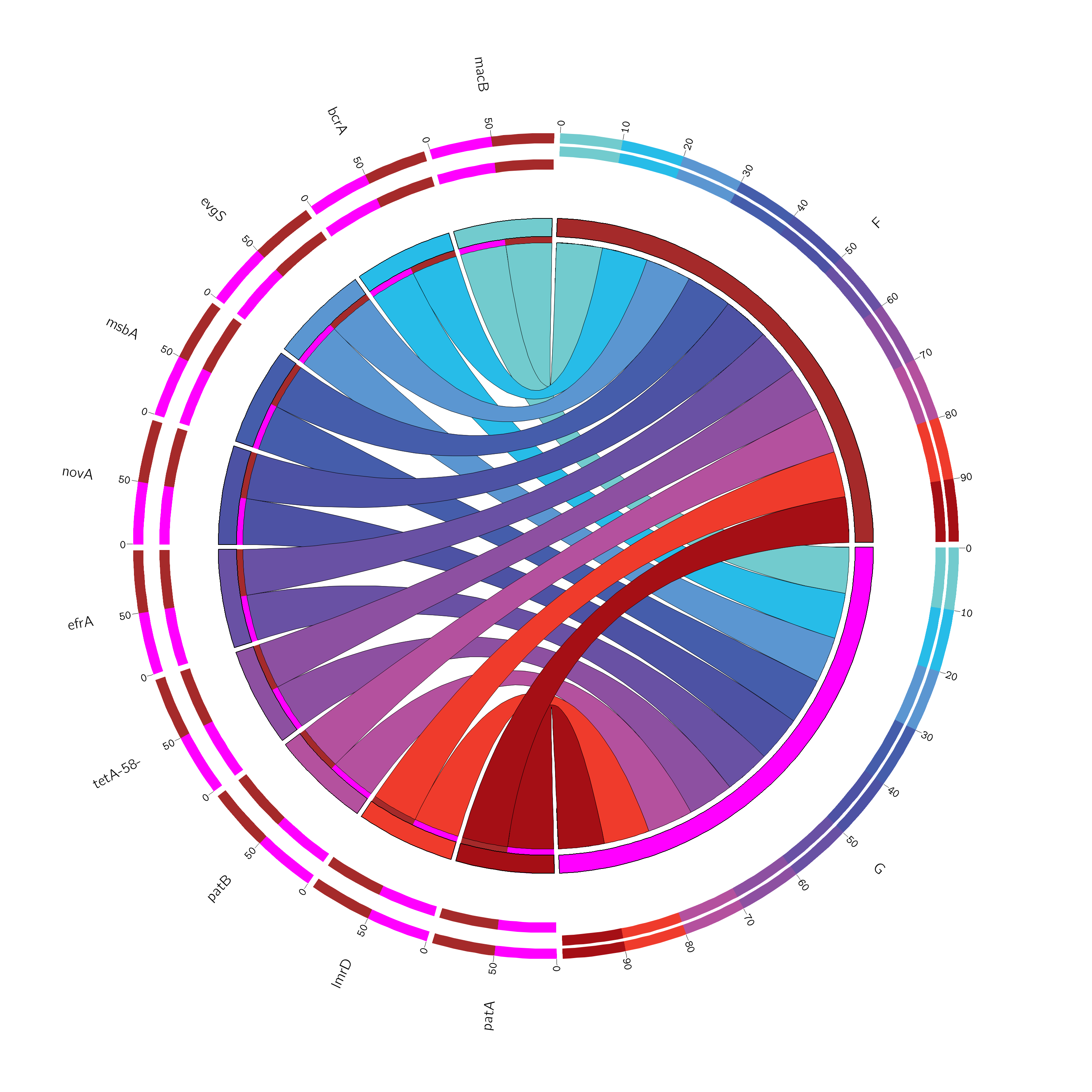
6.6 PHI注释
PHI-base(Pathogen Host Interactions),病原宿主互作数据库,是一个免费开放的数据库,收录了经过实验验证或文献报道的能够感染植物、动物、真菌和昆虫的真菌、卵菌、细菌等病原菌的致病基因、毒力基因和效应蛋白基因。另外,数据库还收录了抗真菌化合物及其靶基因。
最新版本为4.6,共收录6438个基因、11340对互作关系、263个病原菌、194个宿主、510种疾病。数据库包含核酸序列、蛋白序列、功能注释、其他外部数据库(NCBI taxonomy)的注释信息。
我们使用 Diamond 与数据库进行比对注释
Tab 6-6-1 PHI注释表
| GeneID | Subject | PHI-baseAccession | GeneName | Pathogen_NCBI_Taxonomy_ID | PathogenSpecies | Disease | HostDescription | Host_NCBI_Taxonomy_ID | HostSpecies | Phenotype | GeneFunction | F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 |
|---|
| Unigene16 | A0A0D5YKF1 | PHI:8812|PHI:8812 | CopA (ABUW_2707)|CopA (ABUW_2707) | -|- | Acinetobacter baumannii|Acinetobacter baumannii | Nosocomial infections|Nosocomial infections | Rodents|Rodents | 10090|10090 | Mus musculus (related: house mouse)|Mus musculus (related: house mouse) | unaffected pathogenicity|reduced virulence | Copper-translocating P-type ATPase|Copper-translocating P-type ATPase | 0 | 0 | 1.8493628110390513e-07 | 2.4037713864265717e-07 | 0 | 2.967263527620739e-06 | 0 | 0 |
| Unigene44 | I1RCH5 | PHI:1569 | GzOB009 | 449239 | Fusarium graminearum | Fusarium ear blight | Monocots | 4564 | Triticum (related: wheat) | lethal | transcription factor | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene51 | Q5KN74 | PHI:7207 | msh2 | - | Cryptococcus neoformans | Fungal meningitis | Rodents | 10090 | Mus musculus (related: house mouse) | unaffected pathogenicity | DNA mismatch repair protein | 5.625020382828881e-07 | 0 | 0 | 1.9695211668821954e-07 | 0 | 4.630888308713168e-07 | 0 | 0 |
| Unigene60 | Q8ZQE4 | PHI:3928 | MacB | 1299114 | Salmonella enterica | Food poisoning; enteritis | Rodents | 10090 | Mus musculus (related: house mouse) | reduced virulence | ABC-Type Efflux Pump | 3.556833891525611e-07 | 3.192848354869383e-07 | 3.85283918966469e-07 | 0 | 0 | 0 | 1.222359715024187e-07 | 0 |
| Unigene63 | A6QHK8 | PHI:3004 | ClpX | 93061 | Staphylococcus aureus | Skin infections; food poisoning; respiratory diseases | Rodents | 10090 | Mus musculus (related: house mouse) | reduced virulence | Part of proteolytic complex | 9.383718213376012e-07 | 0 | 1.7425107375123505e-07 | 4.246662782686943e-07 | 0 | 0 | 0 | 0 |
| Unigene68 | Q839C1 | PHI:4139|PHI:4139 | ldh-1|ldh-1 | 226185|226185 | Enterococcus faecalis|Enterococcus faecalis | Nosocomial infections|Nosocomial infections | Rodents|Rodents | 10090|10090 | Mus musculus (related: house mouse)|Mus musculus (related: house mouse) | unaffected pathogenicity|reduced virulence | Redox Balance|Redox Balance | 0 | 0 | 4.415145449777915e-07 | 0 | 0 | 0 | 0 | 0 |
| Unigene144 | Q8DNW9 | PHI:6096 | PepO | 373153 | Streptococcus pneumoniae | Pneumococcal pneumonia | Rodents | 10090 | Mus musculus (related: house mouse) | increased virulence (hypervirulence) | Endopeptidase | 2.4816795455809075e-05 | 2.1444175455707123e-05 | 1.8403891096352753e-05 | 1.0683850988088986e-05 | 3.628834343687883e-05 | 6.83472219294583e-05 | 9.362398618977449e-05 | 2.465023983293573e-06 |
| Unigene148 | G4NDE1 | PHI:1017|PHI:2067 | ABC4|ABC4 | -|- | Magnaporthe oryzae|Magnaporthe oryzae | Rice blast|Rice blast | Monocots|Monocots | 4513|4513 | Hordeum vulgare (related: barley)|Hordeum vulgare (related: barley) | reduced virulence|loss of pathogenicity | ABC Transporter|multidrug resistance | 0 | 5.455598110104968e-07 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene154 | Q882L6 | PHI:3350 | bifA | 223283 | Pseudomonas syringae | Bacterial speck | Eudicots | 4081 | Solanum lycopersicum (related: tomato) | reduced virulence | c-di-GMP phosphodiesterase | 0 | 5.228455178859191e-07 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unigene171 | A4VXN7 | PHI:6897 | 1910RR (ssu05 1910) | 391295 | Streptococcus suis | Meningitis | Even-toed ungulates | 9823 | Sus scrofa (related: pig) | reduced virulence | virulence related two-component system | 0 | 2.6159762366466053e-07 | 0 | 3.461953355451312e-07 | 0 | 0 | 0 | 0 |
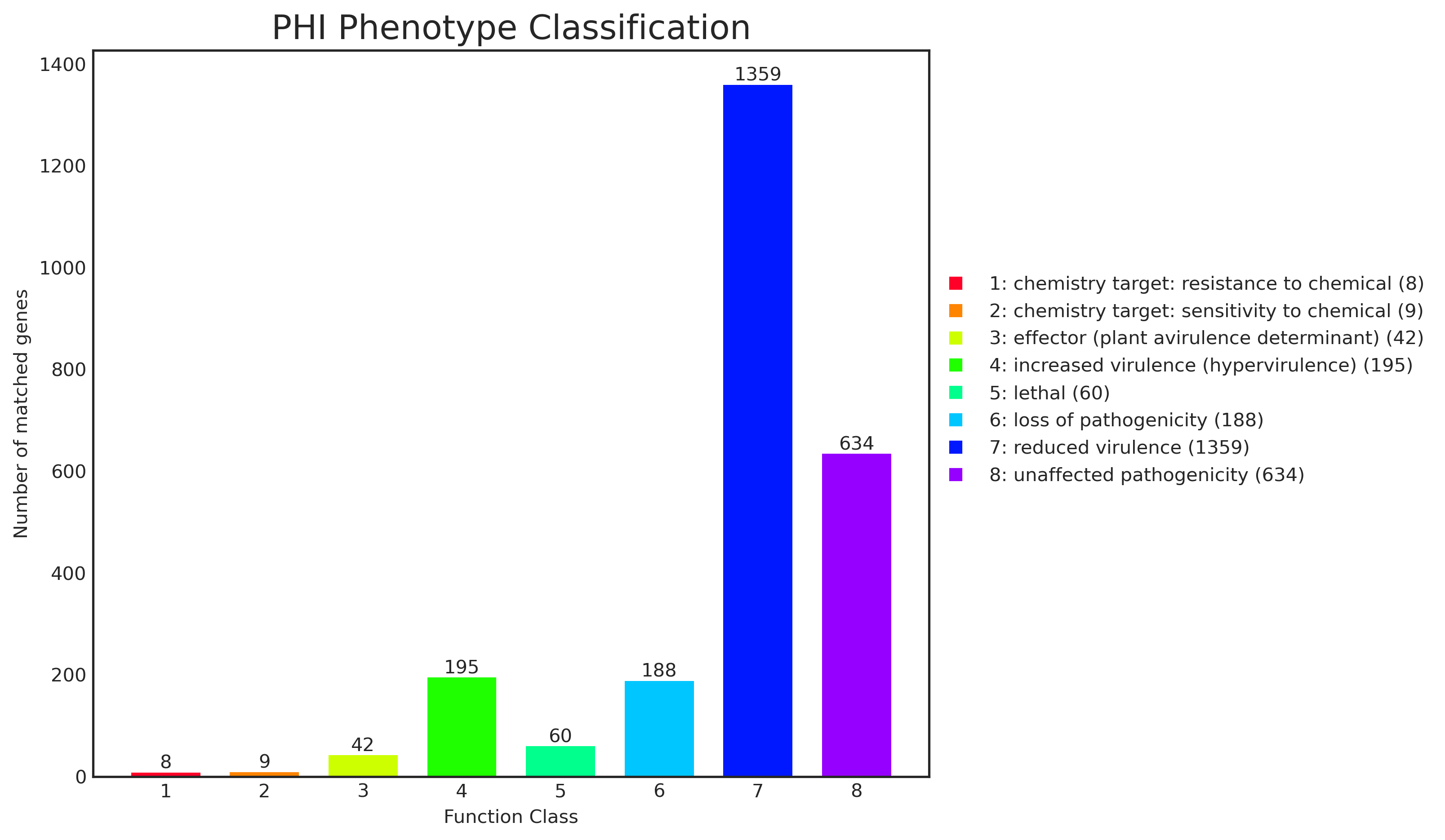
Tab 6-6-2 PHI 数据库表型信息统计
| Phenotype Classificatin | chemistry target: resistance to chemical | chemistry target: sensitivity to chemical | effector (plant avirulence determinant) | increased virulence (hypervirulence) | lethal | loss of pathogenicity | reduced virulence | unaffected pathogenicity |
|---|
| Num | 8 | 9 | 42 | 195 | 60 | 188 | 1359 | 634 |
![]() |
Fig 6-6-1 表型分类统计柱形图 |
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
7 物种组成分析
除了丰富特色的功能分析,宏基因组的另一大特色就是将物种研究推向了更精细的层面。相对于16S,宏基因组可以获得更丰富的属、种水平物种,因而通常用于对16S的拓展补充。基于各层级的物种丰度表,可以结合堆叠图、Circos图、热图等进行物种分布特征的初步展示,即物种组成分析。
7.1 物种注释
当前主流的物种注释有两种方法,即基于reads或基于基因。不同方法各有特色,在文章中也都有广泛应用:
1)基于reads进行物种注释:不受样本复杂度、组装结果的影响,在物种定性定量研究中更准确,使用率较高。
方法:使用Kaiju软件将reads比对Nr微生物库(包含细菌、真菌、古细菌、病毒、微小动植物)进行物种注释。
2)基于基因进行物种注释:受样本、组装影响,定性定量不一定准确,但可基于数据库找到功能-物种的对应关系,用于特定功能的物种分析。
方法:使用DIOMAND软件将非冗余基因集的unigene序列比对Nr微生物库,使用MEGAN(MEtaGenome Analyzer)软件的LCA算法,获得物种注释信息。
注:我们提供两种注释方案可选,若无选择,则默认第一种基于reads注释的方法。两种注释方法详细的比较见
各层级物种总注释信息统计如下:
Tab 7-1-1 物种注释总表
| ID | F-3_tagNumber | F-4_tagNumber | F-5_tagNumber | F-6_tagNumber | G-2_tagNumber | G-4_tagNumber | G-5_tagNumber | G-6_tagNumber | F-3_relativeAbundance | F-4_relativeAbundance | F-5_relativeAbundance | F-6_relativeAbundance | G-2_relativeAbundance | G-4_relativeAbundance | G-5_relativeAbundance | G-6_relativeAbundance |
|---|
| r__root | 19583451.0 | 23774457.0 | 21561531.0 | 21978713.0 | 24193302.0 | 21887107.0 | 21640329.0 | 23204817.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| r__root|k__Archaea | 156995.0 | 190165.0 | 136392.0 | 190712.0 | 464969.0 | 196153.0 | 103382.0 | 664393.0 | 0.00801671778891269 | 0.007998710548888666 | 0.006325710358879432 | 0.008677123178231591 | 0.019218914392090836 | 0.008962034132697392 | 0.004777284116151839 | 0.02863168453343114 |
| r__root|k__Archaea|p__Archaea_noname | 1666.0 | 2063.0 | 1676.0 | 2858.0 | 1762.0 | 1716.0 | 1811.0 | 2415.0 | 8.507182927054072e-05 | 8.677380097471837e-05 | 7.773102939675295e-05 | 0.00013003491150732983 | 7.283007503481749e-05 | 7.840232151284315e-05 | 8.368634321594649e-05 | 0.00010407321893553395 |
| r__root|k__Archaea|p__Archaea_noname|c__Archaea_noname | 1666.0 | 2063.0 | 1676.0 | 2858.0 | 1762.0 | 1716.0 | 1811.0 | 2415.0 | 8.507182927054072e-05 | 8.677380097471837e-05 | 7.773102939675295e-05 | 0.00013003491150732983 | 7.283007503481749e-05 | 7.840232151284315e-05 | 8.368634321594649e-05 | 0.00010407321893553395 |
| r__root|k__Archaea|p__Archaea_noname|c__Archaea_noname|o__Archaea_noname | 1666.0 | 2063.0 | 1676.0 | 2858.0 | 1762.0 | 1716.0 | 1811.0 | 2415.0 | 8.507182927054072e-05 | 8.677380097471837e-05 | 7.773102939675295e-05 | 0.00013003491150732983 | 7.283007503481749e-05 | 7.840232151284315e-05 | 8.368634321594649e-05 | 0.00010407321893553395 |
| r__root|k__Archaea|p__Archaea_noname|c__Archaea_noname|o__Archaea_noname|f__Archaea_noname | 1666.0 | 2063.0 | 1676.0 | 2858.0 | 1762.0 | 1716.0 | 1811.0 | 2415.0 | 8.507182927054072e-05 | 8.677380097471837e-05 | 7.773102939675295e-05 | 0.00013003491150732983 | 7.283007503481749e-05 | 7.840232151284315e-05 | 8.368634321594649e-05 | 0.00010407321893553395 |
| r__root|k__Archaea|p__Archaea_noname|c__Archaea_noname|o__Archaea_noname|f__Archaea_noname|g__Archaea_noname | 1666.0 | 2063.0 | 1676.0 | 2858.0 | 1762.0 | 1716.0 | 1811.0 | 2415.0 | 8.507182927054072e-05 | 8.677380097471837e-05 | 7.773102939675295e-05 | 0.00013003491150732983 | 7.283007503481749e-05 | 7.840232151284315e-05 | 8.368634321594649e-05 | 0.00010407321893553395 |
| r__root|k__Archaea|p__Archaea_noname|c__Archaea_noname|o__Archaea_noname|f__Archaea_noname|g__Archaea_noname|s__Candidatus_Geothermarchaeota_archaeon | 33.0 | 106.0 | 52.0 | 157.0 | 27.0 | 51.0 | 66.0 | 76.0 | 1.6850962580599303e-06 | 4.4585666036452486e-06 | 2.4117025827154853e-06 | 7.143275404706363e-06 | 1.1160113654597458e-06 | 2.330138926081003e-06 | 3.049861210520413e-06 | 3.2751820451762235e-06 |
| r__root|k__Archaea|p__Archaea_noname|c__Archaea_noname|o__Archaea_noname|f__Archaea_noname|g__Archaea_noname|s__Candidatus_Geothermarchaeota_archaeon_ex4572_27 | 30.0 | 26.0 | 38.0 | 22.0 | 38.0 | 31.0 | 29.0 | 21.0 | 1.5319056891453912e-06 | 1.0936106763658157e-06 | 1.7623980412151623e-06 | 1.0009685280480253e-06 | 1.5706826624989016e-06 | 1.416358955068845e-06 | 1.340090531895333e-06 | 9.049845124829039e-07 |
| r__root|k__Archaea|p__Archaea_noname|c__Archaea_noname|o__Archaea_noname|f__Archaea_noname|g__Archaea_noname|s__Candidatus_Pacearchaeota_archaeon | 169.0 | 245.0 | 202.0 | 216.0 | 188.0 | 155.0 | 167.0 | 148.0 | 8.629735382185703e-06 | 1.0305177527293263e-05 | 9.368536955933231e-06 | 9.82769100265334e-06 | 7.770745803941935e-06 | 7.081794775344224e-06 | 7.71707306298347e-06 | 6.377986087974751e-06 |
使用 KRONA 对物种注释结果 进行可视化展示。圆圈从内到外依次代表不同的分类级别,扇形的大小代表不同OTU注释结果的相对比例。
KRONA可视化结果
7.2 物种丰度统计
优势物种很大程度决定着微生物群落的生态结构以及功能结构,了解群落在各个水平的物种组成情况能有 效地对群落结构的形成、改变以及生态影响等进行解读。
我们统计了各个层级分类水平上的各样品的物种组成情况
结果在文件夹:05.Taxonomy下
Tab 7-2-1 物种丰度表(门水平)
| Level | Phylum | F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 |
|---|
| 2 | Archaea_noname | 8.507182927054072e-05 | 8.677380097471837e-05 | 7.773102939675295e-05 | 0.00013003491150732983 | 7.283007503481749e-05 | 7.840232151284315e-05 | 8.368634321594649e-05 | 0.00010407321893553395 |
| 2 | Candidatus_Aenigmarchaeota | 1.6085009736026607e-05 | 2.2797576407318155e-05 | 2.1890838827725175e-05 | 1.938239058856631e-05 | 1.3474803894069524e-05 | 1.2884297591271428e-05 | 1.4047845575730387e-05 | 1.5169264209237245e-05 |
| 2 | Candidatus_Bathyarchaeota | 0.00017683298005034966 | 0.00014852074224029597 | 0.00017276138693490735 | 0.0001427745109552138 | 0.0001304493284959614 | 0.0001498142262474433 | 0.0001626592645610887 | 0.00013669575588551292 |
| 2 | Candidatus_Diapherotrites | 4.953161728236765e-06 | 6.814035752740851e-06 | 8.162685664575489e-06 | 1.6015496448768405e-05 | 9.42409597499341e-06 | 7.767129753603343e-06 | 4.2513216873920906e-06 | 8.144860612346135e-06 |
| 2 | Candidatus_Heimdallarchaeota | 2.251901363043725e-05 | 2.469036411641284e-05 | 2.1519807661153562e-05 | 4.2586661011497805e-05 | 1.682283799044876e-05 | 2.2753121278202735e-05 | 2.7541170931366154e-05 | 2.3658880826338774e-05 |
| 2 | Candidatus_Korarchaeota | 2.8442382295132762e-05 | 2.7508514705509362e-05 | 2.5090982639405336e-05 | 3.294096428667138e-05 | 1.7690846830250784e-05 | 2.6636686155004404e-05 | 2.647834050951813e-05 | 3.1976119441062604e-05 |
| 2 | Candidatus_Lokiarchaeota | 1.884243997648831e-05 | 2.023179751276759e-05 | 1.2986090830006459e-05 | 1.5151023629090566e-05 | 2.095621341807745e-05 | 1.9098001394154102e-05 | 1.8345377281463698e-05 | 2.5511944351899004e-05 |
| 2 | Candidatus_Marsarchaeota | 4.54465354446466e-06 | 8.706823461835532e-06 | 4.962541852895326e-06 | 6.506295432312165e-06 | 5.745391844403877e-06 | 4.705966850712613e-06 | 5.683832255969861e-06 | 4.007788555281432e-06 |
| 2 | Candidatus_Micrarchaeota | 2.1293489079120936e-05 | 2.5994284538233618e-05 | 2.5322877118512596e-05 | 2.602518172924866e-05 | 2.463491754866698e-05 | 1.955489137966018e-05 | 2.5184459995964017e-05 | 2.417601483347186e-05 |
| 2 | Candidatus_Odinarchaeota | 1.5829692121169041e-06 | 1.2197965236387943e-06 | 1.2522301871791942e-06 | 1.8654413477258654e-06 | 1.446681399670041e-06 | 2.83271791013769e-06 | 1.1552504585304595e-06 | 2.1547250297212e-06 |
7.3 物种分布堆叠图
我们使用堆叠图直观展示各个层级分类水平上各样品的物种丰度情况,初步呈现样本间的物种分布规律,优势物种等信息。由于在堆叠图中无法呈现丰度过低的物种,我们只展示总丰度排名前10的高丰度物种,其余物种统一归类到Other类别,无法注释到该水平的序列则被归类到Unclassified类别。
- Kingdom 水平物种分布堆叠图
- Phylum 水平物种分布堆叠图
- Class 水平物种分布堆叠图
- Order 水平物种分布堆叠图
- Family 水平物种分布堆叠图
- Genus 水平物种分布堆叠图
- Species 水平物种分布堆叠图
Fig 7-3-1 各分类水平物种分布堆叠图
7.4 物种分布热图
热图比较美观,可以包含的信息量相对较多,我们使用热图展示更多物种的丰度信息。挑选物种总丰度排名前20的物种,使用R语言pheatmap包绘制热图,详细展示各个层级分类水平上各样品的物种丰度高低。
- Kingdom 水平物种分布热图
- Phylum 水平物种分布热图
- Class 水平物种分布热图
- Order 水平物种分布热图
- Family 水平物种分布热图
- Genus 水平物种分布热图
- Species 水平物种分布热图
Fig 7-4-1 各分类水平物种分布热图
7.5 物种分布Circos图
Circos图以更新颖的形式展示物种丰度分布规律,通过图中连线的粗细展示各样本的优势物种。对各分类水平,我们使用Circos软件绘制Circos图展示前10个高丰度物种。
- Phylum 水平分布Circos图
- Class 水平分布Circos图
- Order 水平分布Circos图
- Family 水平分布Circos图
- Genus 水平分布Circos图
- Species 水平分布Circos图
Fig 7-5-1 各分类水平物种分布Circos图
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
8 Alpha 多样性分析
8.1 Alpha 多样性指数
Alpha多样性是指特定生境或者生态系统内物种/功能的丰富程度,它可以指示生境的平衡状态、生存条件情况等。如有研究发现疾病组肠道菌群失衡,alpha多样性会显著下降。
通常基于物种/功能丰富度(种类的数量)和物种均匀度(丰度高低的均匀程度,若所有物种丰度相等,则均匀度最高)两个指标来评估。
我们主要展示Chao1,ACE(Alternating Conditional Expectation),Shannon,Simpson,四个指数以及相关分析结果。
- Chao1、ACE 指数只考虑物种的丰富度,数值越大,多样性越高。
- Shannon、Simpson综合体现物种的丰富度和均匀度,数值越大,样本的均衡性越高。
Tab 8-1-1 Alpha 多样性统计总表
| Type | Index | F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 |
|---|
| genus | chao1 | 4284.578947368421 | 4285.682692307692 | 4350.74358974359 | 4344.727272727273 | 4337.591397849463 | 4386.754237288134 | 4327.291666666667 | 4378.028301886792 |
| genus | ace | 4237.38392536951 | 4286.720664695999 | 4298.904665243468 | 4351.666340910032 | 4300.745848893888 | 4382.132329593906 | 4318.209889124698 | 4363.759557342247 |
| genus | shannon | 6.699152845288879 | 6.575547858810985 | 6.655635045765983 | 7.1477455237312135 | 6.311606947751441 | 6.573471267449445 | 6.487134516932295 | 6.393029425070315 |
| genus | simpson | 0.9638712273074456 | 0.9588640737838152 | 0.9493260808104316 | 0.9679361442961336 | 0.9385281344544636 | 0.9523271908630014 | 0.9505495465319259 | 0.9321293514407792 |
| species | chao1 | 29398.030769230765 | 29583.772990886493 | 29690.513951395136 | 29927.55117449664 | 29489.01357827476 | 30116.93691588785 | 29320.260517799354 | 29883.00240577386 |
| species | ace | 29027.51448667658 | 29271.553097824813 | 29216.397691246315 | 29531.715458762505 | 29146.07785751545 | 29777.323074345568 | 29062.58592878999 | 29547.499923863794 |
| species | shannon | 9.995232143511453 | 10.035844513037732 | 10.25337058129983 | 10.387318230289695 | 9.963000943598646 | 10.291785756402321 | 9.951717669316853 | 10.2047790112494 |
| species | simpson | 0.9891912973704516 | 0.9891517025446755 | 0.991119970220598 | 0.9908629856663936 | 0.9918034412008476 | 0.9885140675928804 | 0.9854163097255156 | 0.9916189989952692 |
| gene | chao1 | 461191.0348557693 | 555474.2394576988 | 400383.22291904216 | 395696.3141057934 | 388747.06076941924 | 293558.0 | 434788.34883833263 | 588608.6996212524 |
| gene | ace | 461195.7547310578 | 554887.635305827 | 400394.5814852618 | 395710.7460666684 | 388752.04222555825 | 293558.0 | 433940.0586241977 | 587936.1538772129 |
8.2 Alpha 多样性差异分析
不同生境下的环境驱动因素能引起微生物α多样性差异。结合分组和采样信息,通过对两组或者多组间的α多样性进行假设检验,可以分析组间的物种/功能多样性是否存在显著的差异,从而初步判断驱动群落多样性变化的潜在因素等。我们同时使用以下几种主流的假设检验方法进行差异分析。
- 针对2个分组的比较时,使用weltch’s T-test检验和wilcox秩和检验
- 针对2个以上分组的比较时,使用Kruskal-Wallis秩和检验和Tukey检验
(上述的检验方法均要求每组至少含有3个重复样本)
以属水平Chao1指数为例,基于welch’s t 检验、Tukey HSD检验结果用盒型图展示两组差异如下:
结果在文件夹:06.Alpha 下
Fig 8-2-1 Chao1指数Welch’s T检验盒型图
9 Beta 多样性分析
Beta 多样性分析可以用来分析样本关系,评估样本的聚类特征与预期分组是否一致,分组是否显著。我们从基因丰度、基因功能(默认KEGG、eggNOG、CAZy丰度表)以及物种丰度多个角度开展样本关系分析,进行组间差异及组间多元统计等分析,并使用韦恩图展示样本间共有、特有的信息。
9.1 样本相关性分析
同时,为了比较样本(分组)之间的相似性,我们基于物种(基因、功能)计算样本之间的Pearson系数,并进行热图展示。
结果在文件夹:07.Beta/Heatmap下
- gene
- Genus
- kegg.A
- kegg.B
- CAZy.A
- CAZy.B
- eggNOG.A
- eggNOG.C
Fig 9-1-1 样本相关性分析热图
9.2 PCA主成分分析
基于物种(基因、功能)丰度表,可以开展主成分分析(PCA,Principal Component Analysis),从而利用降维的思想研究样本间的距离关系。这种方法借用方差分解可以有效的找出数据中最“主要”的元素和结构,将复杂的样本组成关系反映到横纵坐标的两个特征值上,从而达到简化数据复杂度的效果。分析结果中,样品组成越相似,反映在PCA 图中的距离越近,而且不同环境间的样品往往可能表现出各自聚集的分布情况。
结果在文件夹:07.Beta/PCA下
- gene
- Genus
- kegg.A
- kegg.B
- CAZy.A
- CAZy.B
- eggNOG.A
- eggNOG.C
Fig 9-2-1 PCA分析图
9.3 PCoA主坐标分析
PCoA主坐标分析是一种展示样本间相似性的分析方式,它的分析思路与PCA分析基本一致,都是通过降维方式寻找复杂样本中的主要样本差异距离。与PCA不同的是,PCoA主要利用Bray-Curtis等信息,因此结果更集中于体现样本间结构的相异性。
基于物种(基因、功能)丰度信息,使用R语言计算样本间的Bray-Curtis矩阵,我们可以绘制PCoA图形。分析结果中,样品越相似,反映在PCoA 图中的距离越近,而且不同环境间的样品往往可能表现出各自聚集的分布情况。
结果在文件夹:07.Beta/PCoA下
- gene
- Genus
- kegg.A
- kegg.B
- CAZy.A
- CAZy.B
- eggNOG.A
- eggNOG.C
Fig 9-3-1 PCoA分析图
9.4 NMDS分析
NMDS是非线性模型,适用于无法获得研究对象间精确的相似性或相异性数据,其设计目的是为了克服线性模型(包括PCA、 PCoA)的缺点,更好地反映生态学数据的非线性结构。我们根据PCoA分析所获得的Bray-Curtis矩阵,进行NMDS分析。其特点是根据样品中包含的物种(基因、功能)信息,以点的形式反映在多维空间上,而对不同样品间的差异程度,则是通过点与点间的距离体现的,最终获得样品的空间定位点图。
结果在文件夹:07.Beta/NMDS下
- gene
- Genus
- kegg.A
- kegg.B
- CAZy.A
- CAZy.B
- eggNOG.A
- eggNOG.C
Fig 9-4-1 NMDS分析图
9.5 UPGMA分类树
在微生物生态研究当中,UPGMA分类树可以用于研究样本间的相似性,解答样本的分类学问题。利用R语言,根据物种(基因、功能)所计算的Bray-Curtis矩阵信息,可以将样本进行UPGMA分类树分类。其中越相似的样本将拥有越短的共同分支。
结果在文件夹:07.Beta/UPGMA 下
- Phylum
- Class
- Order
- Family
- Genus
- Species
Fig 9-5-1 UPGMA样本分类树
9.6 Anosim差异检验
Analysis of Similarity (ANOSIM)分析是一种对微生物群落结构的非参数检验方法,用来检验组间的差异是否显著大于组内差异,从而判断分组是否有意义。
根据物种(基因、功能)所计算的Bray-Curtis矩阵信息,我们开展Anosim分析,以属水平为例展示如下。
结果在文件夹:
07.Beta/Anosim_and_Adonis/Anosim 下
Tab 9-6-1 物种水平Anosim分析结果表
| diffs | Rvalue | Pvalue | significant |
|---|
| F-VS-G | -0.0521 | 0.583 | |
根据分组信息,基于Bray-Curtis距离,利用Mothur软件可以计算两两样本之间距离的轶(Ranks),通过比较组内和组间的轶均值,从而获得分组差异信息。
结合Anosim检验结果,我们基于样本间Bray-Curtis距离的秩(rank),使用盒形图展示检验结果。
Fig 9-6-1 物种水平Anosim结果盒形图
9.7 Adonis分析
Adonis是一种基于Bray-Curtis距离矩阵的非参数多元方差分析方法。该方法可分析不同分组因素对样品差异的解释度,并使用置换检验对分组的统计学意义进行显著性分析。根据物种(基因、功能)所计算的Bray-Curtis矩阵信息,我们开展Asonis分析,以属水平示例如下。
结果在文件夹:
06.Comparison/Anosim_and_Adonis/Adonis 下
Tab 9-7-1 基于Bray -Curtis距离的Adonis分析(物种水平)
| diffs | Df | SumsOfSqs | MeanSqs | Fvalue | R2 | Pvalue | significant |
|---|
| F-VS-G | 1 | 0.0189 | 0.0189 | 0.7947 | 0.117 | 0.551 | |
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
10 常规差异分析
10.1 物种韦恩分析
不同生境下的微生物群落,其物种分布存在一定程度的相似性和特异性。
在多分组(或样本)情况下,为了解不同分组(或样本)之间的物种差异情况,我们基于样本的物种丰度信息开展韦恩图(4组以内)/花瓣图(5组以上)和Upset图分析,以展示不同样本之间的共有特有信息,以种水平展示如下:
结果在文件夹:08.Different/Venn/下
Fig 10-1-1 各比较组 种 水平韦恩图(花瓣图)展示
10.2 功能韦恩图分析
类似的,我们也可以统计分组共有、特有的功能特征gene、KEGG、eggNOG、CAZy,辅助了解分组之间的功能差异。
以kegg.B水平示例如下:
Fig 10-2-1 各比较组 kegg.B 水平韦恩图(花瓣图)展示
10.3 物种差异 Welch’s t 检验
使用Welch's T检验(R语言)进行两个分组间的物种(门到种水平,保留在至少一个样本的相对丰度达到0.1%以上的物种参与分析)差异分析,一般以P-value < 0.05(或0.01)为显著性阈值,P-value越小说明物种差异越显著.
Tab 10-3-1 种水平 F_vs_G Welch’s T检验统计
| labels | F | G | fold(G/F) | p-value | significant |
|---|
| Methanobrevibacter_millerae | 0.00143635 | 0.0015162 | 1.05559479 | 0.927357199978657 | no |
| Methanobrevibacter_olleyae | 0.0002818 | 0.00088716 | 3.14822993 | 0.29134122619155 | no |
| Methanobrevibacter_ruminantium | 0.00029661 | 0.00126429 | 4.26250422 | 0.23977526587141 | no |
| Methanobrevibacter_sp._YE315 | 0.00033607 | 0.00173199 | 5.15374671 | 0.181108438782593 | no |
| Methanobrevibacter_thaueri | 0.00055569 | 0.0020649 | 3.71590199 | 0.177588593993751 | no |
| Acidobacteria_bacterium | 0.00118907 | 0.00105044 | 0.88341273 | 0.218483551673163 | no |
| Coriobacteriales_bacterium_OH1046 | 0.00091 | 0.00121596 | 1.33621276 | 0.242712453629302 | no |
| Slackia_heliotrinireducens | 0.00082083 | 0.00099416 | 1.21116807 | 0.333886284437545 | no |
| bacterium_F083 | 0.00366297 | 0.00374867 | 1.02339744 | 0.960365274125692 | no |
| bacterium_P201 | 0.0017276 | 0.00183194 | 1.06039478 | 0.816154555106317 | no |
针对比较组间有显著差异(P< 0.05)的物种,我们使用柱形图直观展示其在两个分组中的丰度和差异显著性。(组间无显著差异的物种,则无图形)。
- Phylum
- Class
- Order
- Family
- Genus
- Species
Fig 10-3-1 F_vs_G 各分类水平差异分析柱状图
10.4 功能差异 Welch’s t 检验
使用 Welch's T检验(R语言)进行两个分组功能(Gene,KEGG,CAZy,eggNOG各功能层级,取比较组中相对丰度之和在top200的功能参与分析)差异分析,结果以P-value < 0.05(或0.01)为阈值,P-value越小说明差异越显著。
Tab 10-4-1 KEGG LevelB F_vs_G Welch’s T检验统计
| labels | F | G | fold(G/F) | p-value | significant |
|---|
| Carbohydrate metabolism | 0.04647864 | 0.04728229 | 1.01729071 | 0.735713042352088 | no |
| Amino acid metabolism | 0.03457677 | 0.03458282 | 1.00017522 | 0.996699325734872 | no |
| Metabolism of cofactors and vitamins | 0.02541726 | 0.0261729 | 1.02972915 | 0.655394569910569 | no |
| Nucleotide metabolism | 0.0240265 | 0.02453778 | 1.02128002 | 0.57338365259418 | no |
| Membrane transport | 0.02321069 | 0.02177247 | 0.93803621 | 0.643425047020106 | no |
| Replication and repair | 0.01919478 | 0.01852055 | 0.96487407 | 0.405822351877779 | no |
| Translation | 0.01800969 | 0.01603198 | 0.8901863 | 0.184060083149889 | no |
| Energy metabolism | 0.01457296 | 0.01459341 | 1.00140333 | 0.973348053832285 | no |
| Signal transduction | 0.01358768 | 0.01404621 | 1.03374597 | 0.449599716284469 | no |
| Cellular community - prokaryotes | 0.01238952 | 0.01218747 | 0.98369156 | 0.896197491569113 | no |
针对比较组间有显著差异(P< 0.05)的功能,我们使用柱形图直观展示其在两个分组中的丰度和差异显著性。(组间无显著差异的功能,则无图形)。
- Gene
- KEGG_A
- KEGG_B
- CAZy_A
- CAZy_B
- eggNOG_A
- eggNOG_C
Fig 10-4-1 F_vs_G 功能差异分析柱状图
10.5 物种差异方差分析
使用方差分析(R语言)进行两个分组间的物种(门到种水平,保留在至少一个样本的相对丰度达到0.1%以上的物种参与分析)差异分析,一般以P-value < 0.05(或0.01)为显著性阈值,P-value越小说明物种差异越显著。
Tab 10-5-1 种水平 F-VS-G 物种 ANOVA 分析
| Species | F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 | p_value | q_value |
|---|
| Methanobrevibacter_millerae | 0.00120147363199673 | 0.00183932697180003 | 0.000912412017495418 | 0.00179218865090053 | 0.00144341603308221 | 0.000597063833059344 | 0.000276751799845557 | 0.0037475839606923 | 0.927357372373287 | 0.968880836807912 |
| Methanobrevibacter_olleyae | 4.83571562540229e-05 | 0.000509917008830107 | 5.49126126525987e-05 | 0.000513997339152661 | 0.0018628296377237 | 0.000136838550659071 | 3.35484733157245e-05 | 0.00151541811340292 | 0.291341205439381 | 0.739004008838848 |
| Methanobrevibacter_ruminantium | 7.99144134504179e-05 | 0.000536626346502887 | 8.47342426657921e-05 | 0.000485151246117095 | 0.00227162046751617 | 0.000200711770632821 | 4.99530298268571e-05 | 0.00253486161946461 | 0.23977526036118 | 0.739004008838848 |
| Methanobrevibacter_sp._YE315 | 0.000547298839208677 | 0.000234537428131376 | 0.000288523110905251 | 0.000273901388129505 | 0.00155716652485056 | 0.00117900460759844 | 0.000202076410206148 | 0.00398973195953237 | 0.181108375728516 | 0.739004008838848 |
| Methanobrevibacter_thaueri | 0.00126576260741787 | 0.000265368836815074 | 0.000345893805036386 | 0.000345743629301679 | 0.00234271452487139 | 0.00130346143965029 | 0.000291123115549676 | 0.00432229222061954 | 0.177588623174531 | 0.739004008838848 |
| Acidobacteria_bacterium | 0.00112191666320711 | 0.00101339853944929 | 0.0012710600188827 | 0.00134989705721168 | 0.000852384680685588 | 0.00114304736573911 | 0.00111745066352734 | 0.00108886874651931 | 0.218482791398186 | 0.739004008838848 |
| Coriobacteriales_bacterium_OH1046 | 0.0010185130291898 | 0.00109756449958037 | 0.000657374469373255 | 0.00086656575387285 | 0.00175746989807344 | 0.00119408197712014 | 0.00114494562444037 | 0.000767340677584314 | 0.242712112480166 | 0.739004008838848 |
| Slackia_heliotrinireducens | 0.000920828509745295 | 0.0010614753472603 | 0.000676760847826622 | 0.000624240372946314 | 0.00134917507333228 | 0.000834326802532651 | 0.00101075173117747 | 0.000782380658291768 | 0.333885745820546 | 0.739004008838848 |
| bacterium_F083 | 0.00229642875507488 | 0.00193308305632385 | 0.00367970159447397 | 0.00674266050063987 | 0.0022082971559649 | 0.00606055427974104 | 0.00107821835795565 | 0.00564762049190045 | 0.960365549960427 | 0.975949493357208 |
| bacterium_P201 | 0.00134327703528862 | 0.00154565044324672 | 0.00177083900025467 | 0.00225063223674653 | 0.00173601767960405 | 0.00232730620817087 | 0.000807566280531132 | 0.00245686057338871 | 0.816154015335662 | 0.892394111597477 |
针对组间有显著差异(P<0.05)的物种或功能,我们使用盒型图展示每个物种在比较组分组当中的丰度,便于直观对比丰度差异。
- Phylum
- Class
- Order
- Family
- Genus
- Species
Fig 10-5-1 F-VS-G 各分类水平差异分析盒型图
10.6 功能差异方差分析
使用方差分析(ANOVA,Analysis of Variance)(R语言)进行两个分组功能(Gene,KEGG,CAZy,eggNOG各功能层级,取比较组中相对丰度之和在top200的功能参与分析)差异分析,结果以P-value < 0.05(或0.01)为阈值,P-value越小说明差异越显著。
Tab 10-6-1 KEGG LevelB F-VS-G 功能 ANOVA 分析
| LevelB | F-3 | F-4 | F-5 | F-6 | G-2 | G-4 | G-5 | G-6 | p_value | q_value |
|---|
| Carbohydrate metabolism | 0.0511722798527289 | 0.0475148219510415 | 0.0430928753325583 | 0.0441345685126972 | 0.0495518564388103 | 0.0475266139576903 | 0.0485761195544872 | 0.0434745508140962 | 0.735713050286961 | 0.873659247215766 |
| Amino acid metabolism | 0.0374200817028052 | 0.034570453182529 | 0.0311621219527577 | 0.0351544063881205 | 0.0358043745875435 | 0.0336712954946209 | 0.0346402109523166 | 0.034215416419643 | 0.996699356971458 | 0.996699356971458 |
| Metabolism of cofactors and vitamins | 0.0255564716807886 | 0.0240512384375626 | 0.0225924835213308 | 0.0294688565575848 | 0.0274328548900081 | 0.0253732482715832 | 0.0254334057336554 | 0.0264520758093085 | 0.655394635990173 | 0.872016482341493 |
| Nucleotide metabolism | 0.0256776156249902 | 0.0246956334907115 | 0.0223287532194466 | 0.0234039950097027 | 0.0238930919942174 | 0.024564405916867 | 0.0257290448664278 | 0.0239645925160805 | 0.5733834885266 | 0.872016482341493 |
| Membrane transport | 0.0286843868223479 | 0.0236866298693737 | 0.0182646588914228 | 0.0222070975317881 | 0.0206041387219408 | 0.0218104327085646 | 0.0271790285691243 | 0.0174962834524385 | 0.643425040157055 | 0.872016482341493 |
| Replication and repair | 0.0196839542631069 | 0.0206427590312089 | 0.0174801444753403 | 0.0189722791929602 | 0.018007311148155 | 0.0190888883997282 | 0.0189771414895626 | 0.0180088576109179 | 0.405822425504613 | 0.872016482341493 |
| Translation | 0.0195916328060651 | 0.0200715031628188 | 0.0155366403043922 | 0.0168389826502198 | 0.0158163105889344 | 0.0145080138600496 | 0.0178249632650667 | 0.0159786288769253 | 0.184060095372788 | 0.872016482341493 |
| Energy metabolism | 0.0154284280514352 | 0.0145218594469978 | 0.0131577186024404 | 0.0151838143431471 | 0.0152549125384274 | 0.013913009991201 | 0.01472341292507 | 0.0144822878080533 | 0.973347665356249 | 0.996699356971458 |
| Signal transduction | 0.0141834815288719 | 0.0135596410702269 | 0.0126581293817935 | 0.0139494568247399 | 0.0148469393661482 | 0.0145652379587045 | 0.0128025922261919 | 0.0139700568239606 | 0.449599919663306 | 0.872016482341493 |
| Cellular community - prokaryotes | 0.0148030563841526 | 0.0123941626535127 | 0.0108704365317802 | 0.0114904366219596 | 0.0109787689533888 | 0.012975702607063 | 0.0151468057773745 | 0.00964859975212261 | 0.896197498434501 | 0.996699356971458 |
针对组间有显著差异(P<0.05)的功能,我们使用盒型图展示每个功能在比较组分组当中的丰度,便于直观对比丰度差异。
- Gene
- KEGG_A
- KEGG_B
- CAZy_A
- CAZy_B
- eggNOG_A
- eggNOG_C
Fig 10-6-1 F-VS-G 功能差异分析盒型图
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
11 个性化差异分析
11.1 物种差异 Metastats 分析
MetaStats 可用于两组间的差异分析。是不同方法的综合。首先进行T检验计算,若组内物种数量少于样本重复数,则基于Fisher精确检验计算P值;若组内物种数大于样本重复数,且重复数大于等于8,则进行单物种的Permutation test 置换检验计算P值,若组内物种数大于样本重复数,且重复数小于8,则混合整个样本基于Permutation test 置换检验计算P值。最后进行多重检验校正计算q值。
结果在文件夹:08.Different/MetaStats下
Tab 11-1-1 Metastats 物种差异分析表(F-VS-G)
| Phylum | Mean(F%) | variance(F%) | std.err(F%) | Mean(G%) | variance(G%) | std.err(G%) | P-value | FDR |
|---|
| Candidatus_Saccharibacteria | 0.0052733151033589 | 3.03066539687542e-06 | 0.000870440319159709 | 0.00213362022078524 | 1.39340840826156e-06 | 0.000590213607150319 | 0.0113846153846154 | 0.142307692307692 |
| Tenericutes | 0.00398534336174878 | 3.6553101405306e-07 | 0.000302295804657069 | 0.00229076609395827 | 4.23786334244343e-07 | 0.000325494367940654 | 0.00634615384615385 | 0.142307692307692 |
| Chlamydiae | 0.00137304119352609 | 9.55323228260157e-08 | 0.000154541517743627 | 0.00105391181338721 | 2.95128132716907e-09 | 2.71628483740617e-05 | 0.0606923076923077 | 0.307692307692308 |
| Fusobacteria | 0.00213099773071195 | 1.83858953638619e-07 | 0.000214393886129373 | 0.00170046818232271 | 7.47403154228998e-09 | 4.32262407059936e-05 | 0.0738461538461539 | 0.307692307692308 |
| Lentisphaerae | 0.00140427233126215 | 1.12295252740029e-07 | 0.000167552419215621 | 0.000968577423638092 | 7.27282894093774e-08 | 0.00013484091497889 | 0.0618076923076923 | 0.307692307692308 |
| Viruses_noname | 0.00169404607277827 | 8.50346249512059e-07 | 0.000461071103386468 | 0.000706842287509728 | 1.5409744808716e-07 | 0.000196276239065736 | 0.0727307692307692 | 0.307692307692308 |
| Verrucomicrobia | 0.0027240930150824 | 2.52278387680304e-07 | 0.000251136610075226 | 0.00212042235083182 | 1.69763488428322e-07 | 0.000206011825163219 | 0.095 | 0.339285714285714 |
| Candidatus_Melainabacteria | 0.000689823051810787 | 1.63949934891477e-07 | 0.000202453658210637 | 0.000369356783064785 | 3.28645845598799e-08 | 9.06429596823161e-05 | 0.1955 | 0.382396449704142 |
| Elusimicrobia | 0.000823831487350264 | 3.47778748478986e-07 | 0.00029486384505352 | 0.000395614643107053 | 3.80477483381041e-09 | 3.08414284437767e-05 | 0.196615384615385 | 0.382396449704142 |
| Euryarchaeota | 0.00734091277024641 | 1.04201495822828e-06 | 0.000510395669610422 | 0.0156475444075367 | 0.000131986630691866 | 0.00574427172694385 | 0.198846153846154 | 0.382396449704142 |
11.2 代谢通路 reporter score 分析
对比较组内pathway,我们基于reporter_score算法,进行精细的差异分析,获得pathway的得分,统计如下
结果在文件夹:08.Different/ReportScore下
Tab 11-2-1 F_vs_G pathway 差异分析 reporter_score
| Pathway ID | Pathway | ReporterScore | Sig | F | G | KO Number | ... |
|---|
| ko00230 | Purine metabolism | 0.038570961581914504 | ns | 0.016964615852905825 | 0.017348519226301748 | 37930 | ... |
| ko02010 | ABC transporters | -1.988922338037288 | * | 0.017147460514868477 | 0.016236825403914214 | 33855 | ... |
| ko00240 | Pyrimidine metabolism | 0.9393183011863767 | ns | 0.01481074029712802 | 0.015269980297308408 | 33616 | ... |
| ko02020 | Two-component system | -0.03572494608188855 | ns | 0.013587677201408056 | 0.014046206593751288 | 27270 | ... |
| ko02024 | Quorum sensing | -0.9630553611131618 | ns | 0.01238952304785125 | 0.01218746927248724 | 24892 | ... |
| ko00520 | Amino sugar and nucleotide sugar metabolism | -0.9956493455424241 | ns | 0.01068069413225684 | 0.010584777998041741 | 24328 | ... |
| ko00500 | Starch and sucrose metabolism | 0.07717280429956615 | ns | 0.01018990239716327 | 0.011673325635221084 | 23047 | ... |
| ko03440 | Homologous recombination | 2.7197871018379054 | * | 0.00988389172238257 | 0.009256773657304618 | 20545 | ... |
| ko00970 | Aminoacyl-tRNA biosynthesis | 3.2822402463154177 | * | 0.008881198487858056 | 0.008603023366557166 | 20316 | ... |
| ko03430 | Mismatch repair | 1.5135848191578274 | ns | 0.008274938084926976 | 0.0080526204505079 | 17826 | ... |
我们挑选显著富集的20条代谢通路,绘制柱形图展示如下:
Fig 11-2-1 Reporter Score 柱形图
我们挑选显著富集的20条代谢通路,绘制气泡图展示如下:
Fig 11-2-2 Reporter Score 气泡图
11.3 物种差异 LEfSe 分析
通过LDA Effect Size(LefSe)分析组间菌群差异,可以找出各组间特异的主要菌群,有助于开发biomarker等研究。根据物种(基因、功能)在各层级的丰度信息(物种在至少一个样本的相对丰度达到0.1%以上),我们开展LefSe分析。
结果在文件夹:08.Different/LefSe下
11.4 功能差异 LEfSe 分析
类似的,我们可以将LEfSe的分析方法应用于功能差异,分析原理与物种LEfSe分析一致。我们使用LEfSe软件,针对KEGG、eggNOG、CAZy等主要数据库开展高丰度top200功能的差异分析。图形的分枝树,代表不同功能层级,如KEGG数据库,由内至外,对应Level A、B、C。以KEGG示例如下:
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
12 软件版本和参考文献汇总
Tab 12-0-1 软件版本和参考文献
| 分析软件/方法 | 功能 | 版本 | 参考文献 |
|---|
| Fastp | Illumina测序数据校正 | version 0.18.0 | Chen S, Zhou Y, Chen Y, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor[J]. bioRxiv, 2018: 274100. |
| MEGAHIT | reads组装 | version 1.1.2 | Li D, Liu C M, Luo R, et al. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph[J]. Bioinformatics, 2015, 31(10): 1674-1676. |
| MetaGeneMark | 基因预测 | version 3.38 | Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences[J]. Nucleic acids research, 2010, 38(12): e132-e132. |
| CD-HIT | 基因聚类 | version 4.6 | Fu L, Niu B, Zhu Z, et al. CD-HIT: accelerated for clustering the next-generation sequencing data[J]. Bioinformatics, 2012, 28(23): 3150-3152. |
| Bowtie | reads比对contig、基因 | version 2.2.5 | Langmead B, Salzberg S L. Fast gapped-read alignment with Bowtie 2[J]. Nature methods, 2012, 9(4): 357. |
| 基因丰度计算 | 计算公式 | | Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes[J]. Nature, 2012, 490(7418): 55-60. |
| DIAMOND | 基因比对数据库 | version 0.9.24 | Buchfink B, Xie C, Huson D H. Fast and sensitive protein alignment using DIAMOND[J]. Nature methods, 2015, 12(1): 59. |
| kaiju | 基于reads的物种注释 | version 1.6.3 | Menzel P, Ng K L, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju[J]. Nature communications, 2016, 7: 11257. |
| MEGAN | 基于基因物种注释软件MEtaGenome Analyzer | | Huson, D.H., Mitra, S., Ruscheweyh, H.-J., Weber, N., and Schuster, S.C. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res 21, 1552-1560. |
| LCA算法 | 基于基因的物种注释算法 | | Huson, D.H., Auch, A.F., Qi, J., and Schuster, S.C. (2007). MEGAN analysis of metagenomic data. Genome Res 17, 377-386. |
| Python的scikit-bio包 | alpha多样性指数 | version 0.5.6 | http://scikit-bio.org/docs/latest/diversity.html#module-skbio.diversity |
| metastats | 物种biomarker | version 20090414 | White, James Robert, Niranjan Nagarajan, and Mihai Pop. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5.4 (2009): e1000352. |
| LEfSe | 物种biomarker | version 1.0 | Segata, Nicola, et al. Metagenomic biomarker discovery and explanation. Genome biology 12.6 (2011): 1. |
| circos | circos图 | version 0.69-3 | Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics[J]. Genome research, 2009, 19(9): 1639-1645. |
| R语言VennDiagram包 | venn图 | | Chen H, Boutros P C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R[J]. BMC bioinformatics, 2011, 12(1): 35. |
| R语言Vegan包 | Bray距离计算/PCA/PCoA/NMDS/UPGMA/Anosim/Adonis/welch's t检验/方差分析/Tukey HSD | | Oksanen J, Blanchet F G, Kindt R, et al. Vegan: community ecology package. R package version 1.17-4[J]. http://cran. r-project. org>. Acesso em, 2010, 23: 2010. |
| R语言ggplot2包 | PCA/PCoA/NMDS/小提琴图/盒形图 | | Wickham H, Chang W. ggplot2: An implementation of the Grammar of Graphics[J]. R package version 0.7, URL: http://CRAN. R-project. org/package= ggplot2, 2008, 3. |
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
13 数据库汇总
Tab 13-0-1 数据库汇总
| 数据库 | 功能 | 版本 | 注释软件 | 注释参数 | 链接 |
|---|
| KEGG | 京都基因与基因组百科全书 | 20200416 | Diamond | evalue<=1e-5 | http://www.genome.jp/kegg/ |
| eggNOG | 基因功能注释 | 5.0.0 | Diamond | evalue<=1e-5 | http://eggnog5.embl.de/#/app/home |
| PHI | 病原宿主互作数据库 | 4.8 | Diamond | evalue<=1e-5 | http://www.phi-base.org/ |
| VFDB | 细菌毒力因子数据库 | 2020.04.17 | Diamond | evalue<=1e-5 | http://www.mgc.ac.cn/VFs/main.htm |
| CARD | 细菌耐药基因数据库 | 3.0.8 | Diamond | evalue<=1e-5 | http://arpcard.mcmaster.ca |
| CAZy | 碳水化合物酶数据库 | 20190808 | Diamond | evalue<=1e-5 | http://www.mgc.ac.cn/VFs/main.htm |
| Nr | 物种注释数据库 | 20190205 | Kaiju | 默认 | https://www.ncbi.nlm.nih.gov/refseq/ |
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
14 目录结构
result 结果目录
├── 01.QC 质控目录
│ ├── 1_Filter_fq/sample/*.new.png(pdf) 过滤后碱基分布图(矢量图)
│ ├── 1_Filter_fq/sample/*.old.png(pdf) 过滤前碱基分布图(矢量图)
│ ├── 1_Filter_fq/filter.stat.fill.png(pdf) 数据预处理分布图(百分比)
│ ├── 1_Filter_fq/filter.stat.count.png(pdf) 数据预处理分布图(数值)
│ ├── 1_Filter_fq/reads_filter.stat.xls 所有样品过滤信息总表
│ ├── 1_Filter_fq/reads_filter.stat.xls 所有样品过滤信息总表
│ ├── 1_Filter_fq/reads_info.stat.xls 所有样品过滤前后碱基质量信息
│ ├── 2_rHost/filter_host.stat.xls 所有样品宿主过滤信息总表
│ └── 2_rHost/filter_host.stack.png(pdf) 所有样本宿主过滤分布图
├── 02.Assemble 组装结果
│ ├── sample/sample.contigs.length_distribution.png(pdf) 各样本组装结果统计位图(矢量图)
│ └── assem.contigs.stat.txt 所有样本组装结果统计
├── 03.Genes 基因预测
│ ├── Unigenes.final.fna(faa) 非冗余基因核酸(蛋白)序列
│ ├── Unigenes.final.gff 非冗余基因gff文件
│ ├── Unigenes.expression.final.xls 非冗余基因表达量总表
│ ├── Unigenes.count.final.xls 非冗余基因counts数
│ ├── Unigenes.abundance.final.xls 非冗余基因丰度
│ ├── bar* 各样本非冗余基因数目柱状图
│ ├── violin* 各分组非冗余基因数目小提琴图
│ └── Core_Pan/* Core-Pan分析结果目录
├── 04.Annotation 基因注释
│ ├── KEGG/* KEGG数据库注释结果
│ ├── eggNOG/* eggNOG数据库注释结果
│ ├── CAZy/* CAZy数据库注释结果
│ ├── CARD/* CARD数据库注释结果
│ ├── VFDB/* VFDB数据库注释结果
│ └── PHI/* PHI数据库注释结果
├── 05.Taxonomy 物种注释
│ ├── profiling.all.xls 各lineage比对丰度信息
│ ├── profiling.all.readnumber.xls 各lineage比对reads number统计
│ ├── profiling.all.relative.xls 各lineage比对相对丰度统计
│ ├── profiling.L*.*.xls 各层级比对相对丰度统计
│ ├── profiling.all.stat.xls 各层级比对上的reads数目统计
│ ├── stack_plot/* 各层级比对信息堆叠图
│ ├── heatmap/* 各层级比对信息热图
│ ├── krona/* krona图形化展示结果
│ └── circos/* 各层级比对记过Circos图展示
├── 06.Comparison 比较分析
│ ├── Venn/* 各层级Venn/UpSet结果
│ ├── Heatmap/* 各层相关性热图结果
│ ├── PCA/* 各层级PCA分析结果
│ ├── PCoA/* 各层级PCoA分析结果
│ ├── NMDS/* 各层级NMDS分析结果
│ ├── UPGMA/* 各层级UPGMA分析结果
│ └── Anosim_and_Adonis/* 各层级Anosim/Adonis分析结果
├── 07.Different 差异分析
│ ├── T_test/* 各层级各比较组Welch’s T分析结果
│ ├── ANOVA/* 各层级各比较组ANOVA分析结果
│ ├── Ternary/* 各层级各比较组三元图分析结果
│ ├── MetaStats/* 各层级各比较组MetaStats分析结果
│ └── LefSe/* 各比较组LefSe分析结果
├── index.html 结题报告
└── src 结果报告内容
├── content.html 结题报告主体
├── css 结题报告js脚本
├── doc 结题报告说明文档
├── image 结题报告图片
└── js 结题报告js脚本
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司
15 附录
15.1 问题解答
15.2 英文方法
15.3 中文实验方法
15.4 引用与致谢
如果您的研究课题使用了基迪奥的测序和分析服务,我们期望您在论文发表时,在Method部分或Acknowledgements部分引用或提及基迪奥公司。
以下语句可供参考:
We are grateful to/thank Guangzhou Genedenovo Biotechnology Co., Ltd for assisting in sequencing and/or
bioinformatics analysis.
 广州基迪奥生物科技有限公司
广州基迪奥生物科技有限公司